Aging hallmarks of the primate ovary revealed by spatiotemporal transcriptomics
- PMID: 38126810
- PMCID: PMC11075111
- DOI: 10.1093/procel/pwad063
Aging hallmarks of the primate ovary revealed by spatiotemporal transcriptomics
Abstract
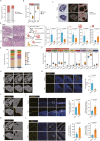
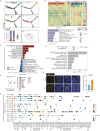
The ovary is indispensable for female reproduction, and its age-dependent functional decline is the primary cause of infertility. However, the molecular basis of ovarian aging in higher vertebrates remains poorly understood. Herein, we apply spatiotemporal transcriptomics to benchmark architecture organization as well as cellular and molecular determinants in young primate ovaries and compare these to aged primate ovaries. From a global view, somatic cells within the non-follicle region undergo more pronounced transcriptional fluctuation relative to those in the follicle region, likely constituting a hostile microenvironment that facilitates ovarian aging. Further, we uncovered that inflammation, the senescent-associated secretory phenotype, senescence, and fibrosis are the likely primary contributors to ovarian aging (PCOA). Of note, we identified spatial co-localization between a PCOA-featured spot and an unappreciated MT2 (Metallothionein 2) highly expressing spot (MT2high) characterized by high levels of inflammation, potentially serving as an aging hotspot in the primate ovary. Moreover, with advanced age, a subpopulation of MT2high accumulates, likely disseminating and amplifying the senescent signal outward. Our study establishes the first primate spatiotemporal transcriptomic atlas, advancing our understanding of mechanistic determinants underpinning primate ovarian aging and unraveling potential biomarkers and therapeutic targets for aging and age-associated human ovarian disorders.
Keywords: aging; inflammation; ovary; primate; senescence; spatial transcriptome.
© The Author(s) 2023. Published by Oxford University Press on behalf of Higher Education Press.
Conflict of interest statement
The authors declare no competing.
Figures





References
Publication types
MeSH terms
Grants and funding
- 2022YFA1103700/National Key Research and Development Program of China
- 81921006/National Natural Science Foundation of China
- YSBR-076/CAS Project for Young Scientists in Basic Research
- FIRMC180305/the Strategic Collaborative Research Program of the Ferring Institute of Reproductive Medicine, Ferring Pharmaceuticals and the Chinese Academy of Sciences
- XDA16000000/the Strategic Priority Research Program of the Chinese Academy of Sciences
- Z190019/the Program of the Beijing Natural Science Foundation
- CAS-WX2021SF-0301/the Informatization Plan of Chinese Academy of Sciences
- 2021-1045/New Cornerstone Science Foundation through the XPLORER PRIZE
- 2022083/Youth Innovation Promotion Association
- 12300927/Excellent Young Talents Program of Capital Medical University
- 11000023T000002036310/the Project for Technology Development of Beijing-affiliated Medical Research Institutes
- BPHR202203105/Excellent Young Talents Training Program for the Construction of Beijing Municipal University Teacher Team
- YESS20210002/Young Elite Scientists Sponsorship Program by CAST
- 2022M712216/The Fellowship of China Postdoctoral Science Foundation
LinkOut - more resources
Full Text Sources
Medical

