Clinical evaluation of a multiplex droplet digital PCR for pathogen detection in critically ill COVID-19 patients with bloodstream infections
- PMID: 38127118
- PMCID: PMC11143000
- DOI: 10.1007/s15010-023-02157-x
Clinical evaluation of a multiplex droplet digital PCR for pathogen detection in critically ill COVID-19 patients with bloodstream infections
Abstract
Background: Nosocomial bloodstream infections (nBSI) have emerged as a clinical concern for physicians treating COVID-19 patients. In this study, we aimed to evaluate the effectiveness of a multiplex ddPCR in detecting bacterial pathogens in the blood of COVID-19 critically ill patients.
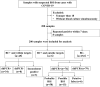
Methods: This prospective diagnostic study included RT-PCR-confirmed COVID-19 patients admitted to our hospital from December 2022 to February 2023. A multiplex ddPCR assay was used to detect common bacterial pathogens and AMR genes in blood samples of the patients, along with antimicrobial susceptibility testing (AST). The diagnostic performance of the ddPCR assay was evaluated by comparing the results with those obtained through blood culture and clinical diagnosis. Additionally, the ability of ddPCR in detecting bacterial resistance was compared with the AST results.
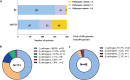
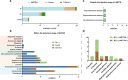
Results: Of the 200 blood samples collected from 184 patients, 45 (22.5%) were positive using blood culture, while 113 (56.5%) were positive for bacterial targets using the ddPCR assay. The ddPCR assay outperformed blood culture in pathogen detection rate, mixed infection detection rate, and fungal detection rate. Acinetobacter baumannii and Klebsiella pneumoniae were the most commonly detected pathogens in COVID-19 critically ill patients, followed by Enterococcus and Streptococcus. Compared to blood culture, ddPCR achieved a sensitivity of 75.5%, specificity of 51.0%, PPV of 30.9%, and NPV of 87.8%, respectively. However, there were significant differences in sensitivity among different bacterial species, where Gram-negative bacteria have the highest sensitivity of 90.3%. When evaluated on the ground of clinical diagnosis, the sensitivity, specificity, PPV and NPV of ddPCR were 78.1%, 90.5%, 94.7%, and 65.5%, respectively. In addition, the ddPCR assay detected 23 cases of blaKPC, which shown a better consistent with clinical test results than other detected AMR genes. Compared to blaKPC, there were few other AMR genes detected, indicating that the application of other AMR gene detection in the COVID-19 critically ill patients was limited.
Conclusion: The multiplex ddPCR assay had a significantly higher pathogen detection positivity than the blood culture, which could be an effective diagnostic tool for BSIs in COVID-19 patients and to improve patient outcomes and reduce the burden of sepsis on the healthcare system, though there is room for optimization of the panels used.- Adjusting the targets to include E. faecalis and E. faecium as well as Candida albicans and Candida glabrata could improve the ddPCR' s effectiveness. However, further research is needed to explore the potential of ddPCR in predicting bacterial resistance through AMR gene detection.
Keywords: Antimicrobial resistance; Bloodstream infections; Multiplex droplet digital PCR; Pathogen detection; Severe Acute Respiratory Syndrome Coronavirus 2.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Clinical evaluation of a multiplex droplet digital PCR for diagnosing suspected bloodstream infections: a prospective study.Front Cell Infect Microbiol. 2025 Jan 16;14:1489792. doi: 10.3389/fcimb.2024.1489792. eCollection 2024. Front Cell Infect Microbiol. 2025. PMID: 39885964 Free PMC article.
-
Clinical validation of a multiplex droplet digital PCR for diagnosing suspected bloodstream infections in ICU practice: a promising diagnostic tool.Crit Care. 2022 Aug 8;26(1):243. doi: 10.1186/s13054-022-04116-8. Crit Care. 2022. PMID: 35941654 Free PMC article.
-
Sensitive and rapid identification of pathogens by droplet digital PCR in a cohort of septic patients: a prospective diagnostic study.Infect Dis (Lond). 2024 Oct;56(10):830-841. doi: 10.1080/23744235.2024.2354312. Epub 2024 May 16. Infect Dis (Lond). 2024. PMID: 38753988
-
Impact of Multiplex PCR on Diagnosis of Bacterial and Fungal Infections and Choice of Appropriate Antimicrobial Therapy.Diagnostics (Basel). 2025 Apr 20;15(8):1044. doi: 10.3390/diagnostics15081044. Diagnostics (Basel). 2025. PMID: 40310414 Free PMC article. Review.
-
The potential impact of the COVID-19 pandemic on global antimicrobial and biocide resistance: an AMR Insights global perspective.JAC Antimicrob Resist. 2021 Apr 8;3(2):dlab038. doi: 10.1093/jacamr/dlab038. eCollection 2021 Jun. JAC Antimicrob Resist. 2021. PMID: 34192258 Free PMC article. Review.
Cited by
-
ddPCR Enhances early diagnosis, treatment, prognosis, and pathogen verification in elderly BSI.Front Cell Infect Microbiol. 2025 Jul 10;15:1605795. doi: 10.3389/fcimb.2025.1605795. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40708754 Free PMC article.
-
Clinical diagnostic performance of droplet digital PCR for pathogen detection in patients with Escherichia coli bloodstream infection: a prospective observational study.BMC Infect Dis. 2025 Jan 6;25(1):22. doi: 10.1186/s12879-024-10396-y. BMC Infect Dis. 2025. PMID: 39757158 Free PMC article.
-
Present and Future Applications of Digital PCR in Infectious Diseases Diagnosis.Diagnostics (Basel). 2024 Apr 29;14(9):931. doi: 10.3390/diagnostics14090931. Diagnostics (Basel). 2024. PMID: 38732345 Free PMC article. Review.
-
Clinical evaluation of a multiplex droplet digital PCR for diagnosing suspected bloodstream infections: a prospective study.Front Cell Infect Microbiol. 2025 Jan 16;14:1489792. doi: 10.3389/fcimb.2024.1489792. eCollection 2024. Front Cell Infect Microbiol. 2025. PMID: 39885964 Free PMC article.
References
-
- WHO. WHO Coronavirus (COVID-19) Dashboard.
-
- Propelled by omicron, U.S. death toll from COVID-19 hits 900,000 Los Angeles, Portland: NewsHour. 2022.
-
- Devi P, Maurya R, Mehta P, Shamim U, Yadav A, Chattopadhyay P, Kanakan A, Khare K, Vasudevan JS, Sahni S, et al. Increased abundance of achromobacter xylosoxidans and bacillus cereus in upper airway transcriptionally active microbiome of COVID-19 mortality patients indicates role of co-infections in disease severity and outcome. Microbiol Spectr. 2022;10:e0231121. doi: 10.1128/spectrum.02311-21. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

