Rab29-dependent asymmetrical activation of leucine-rich repeat kinase 2
- PMID: 38127736
- PMCID: PMC10786121
- DOI: 10.1126/science.adi9926
Rab29-dependent asymmetrical activation of leucine-rich repeat kinase 2
Abstract
Gain-of-function mutations in LRRK2, which encodes the leucine-rich repeat kinase 2 (LRRK2), are the most common genetic cause of late-onset Parkinson's disease. LRRK2 is recruited to membrane organelles and activated by Rab29, a Rab guanosine triphosphatase encoded in the PARK16 locus. We present cryo-electron microscopy structures of Rab29-LRRK2 complexes in three oligomeric states, providing key snapshots during LRRK2 recruitment and activation. Rab29 induces an unexpected tetrameric assembly of LRRK2, formed by two kinase-active central protomers and two kinase-inactive peripheral protomers. The central protomers resemble the active-like state trapped by the type I kinase inhibitor DNL201, a compound that underwent a phase 1 clinical trial. Our work reveals the structural mechanism of LRRK2 spatial regulation and provides insights into LRRK2 inhibitor design for Parkinson's disease treatment.
Conflict of interest statement
Figures

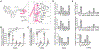

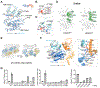

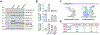
References
-
- Lees AJ, Hardy J, Revesz T, Parkinson’s disease. Lancet 373, 2055–2066 (2009). - PubMed
-
- Tolosa E, Vila M, Klein C, Rascol O, LRRK2 in Parkinson disease: Challenges of clinical trials. Nat. Rev. Neurol 16, 97–107 (2020). - PubMed
-
- Paisán-Ruíz C, Jain S, Evans EW, Gilks WP, Simón J, van derBrug M, López de Munain A, Aparicio S, Gil AM, Khan N, Johnson J, Martinez JR, Nicholl D, Martí Carrera I, Peňa AS, de Silva R, Lees A, Martí-Massó JF, Párez-Tur J, Wood NW, Singleton AB, Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron 44, 595–600 (2004). - PubMed
-
- Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Caine DB, Stoessl AJ, Pfeiffer RF, Patenge N, Carbajal IC, Vieregge P, Asmus F, Müller-Myhsok B, Dickson DW, Meitinger T, Strom TM, Wszolek ZK, Gasser T, Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 44, 601–607 (2004). - PubMed
-
- Ramírez MB, Madero-Perez J, Rivero-Rios P, Martinez-Salvador M, Lara Ordonez AJ, Fernandez B, Fdez E, Hilfiker S, LRRK2 and Parkinson’s disease: From lack of structure to gain of function. Curr. Protein Pept. Sci 18, 677–686 (2017). - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

