Evaluation of anticancer potential of tetracene-5,12-dione (A01) and pyrimidine-2,4-dione (A02) via caspase 3 and lactate dehydrogenase cytotoxicity investigations
- PMID: 38127898
- PMCID: PMC10734984
- DOI: 10.1371/journal.pone.0292455
Evaluation of anticancer potential of tetracene-5,12-dione (A01) and pyrimidine-2,4-dione (A02) via caspase 3 and lactate dehydrogenase cytotoxicity investigations
Abstract
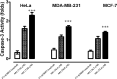
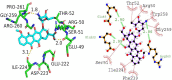
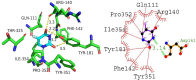
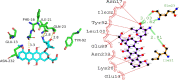
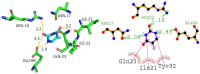
Cancer stands as a significant global cause of mortality, predominantly arising from the dysregulation of key enzymes and DNA. One strategic avenue in developing new anticancer agents involves targeting specific proteins within the cancer pathway. Amidst ongoing efforts to enhance the efficacy of anticancer drugs, a range of crucial medications currently interact with DNA at the molecular level, exerting profound biological effects. Our study is driven by the objective to comprehensively explore the potential of two compounds: (7S,9S)-7-[(2R,4S,5S,6S)-4-amino-5-hydroxy-6-methyloxan-2-yl]oxy-6,9,11-trihydroxy-9-(2-hydroxyacetyl)-4-methoxy-8,10-dihydro-7H-tetracene-5,12-dione (A01) and 5-fluoro-1H-pyrimidine-2,4-dione (A02). These compounds have demonstrated marked efficacy against breast and cervical cancer cell lines, positioning them as promising anticancer candidates. In our investigation, A01 has emerged as a particularly potent candidate, with its potential bolstered by corroborative evidence from lactate dehydrogenase release and caspase-3 activity assays. On the other hand, A02 has exhibited remarkable anticancer potential. To further elucidate their molecular mechanisms and interactions, we employed computational techniques, including molecular docking and molecular dynamics simulations. Notably, our computational analyses suggest that the A01-DNA complex predominantly interacts via the minor groove, imparting significant insights into its mechanism of action. While earlier studies have also highlighted the anticancer activity of A01, our research contributes by providing a deeper understanding of its binding mechanisms through computational investigations. This knowledge holds potential for designing more effective drugs that target cancer-associated proteins. These findings lay a robust groundwork for future inquiries and propose that derivatives of A01 could be synthesized as potent bioactive agents for cancer treatment. By elucidating the distinctive aspects of our study's outcomes, we address the concern of distinguishing our findings from those of prior research.
Copyright: © 2023 Aziz et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures












References
-
- Feng Y, Panwar N, Tng DJH, Tjin SC, Wang K, Yong K-T. The application of mesoporous silica nanoparticle family in cancer theranostics. Coordination chemistry reviews. 2016;319:86–109.
-
- Qi F, Li A, Inagaki Y, Gao J, Li J, Kokudo N, et al. Chinese herbal medicines as adjuvant treatment during chemoor radio-therapy for cancer. Bioscience trends. 2010;4(6). - PubMed
-
- Jabłońska-Trypuć A, Matejczyk M, Rosochacki S. Matrix metalloproteinases (MMPs), the main extracellular matrix (ECM) enzymes in collagen degradation, as a target for anticancer drugs. Journal of enzyme inhibition medicinal chemistry. 2016;31(sup1):177–83. doi: 10.3109/14756366.2016.1161620 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

