The tomato EAR-motif repressor, SlERF36, accelerates growth transitions and reduces plant life cycle by regulating GA levels and responses
- PMID: 38127946
- PMCID: PMC10955490
- DOI: 10.1111/pbi.14228
The tomato EAR-motif repressor, SlERF36, accelerates growth transitions and reduces plant life cycle by regulating GA levels and responses
Abstract
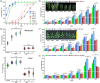
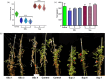
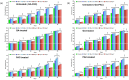
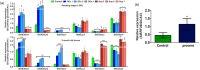
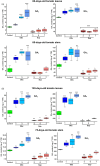
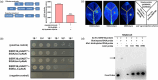
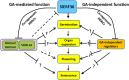
Faster vegetative growth and early maturity/harvest reduce plant life cycle time and are important agricultural traits facilitating early crop rotation. GA is a key hormone governing developmental transitions that determine growth speed in plants. An EAR-motif repressor, SlERF36 that regulates various growth transitions, partly through regulation of the GA pathway and GA levels, was identified in tomato. Suppression of SlERF36 delayed germination, slowed down organ growth and delayed the onset of flowering time, fruit harvest and whole-plant senescence by 10-15 days. Its over-expression promoted faster growth by accelerating all these transitions besides increasing organ expansion and plant height substantially. The plant life cycle and fruit harvest were completed 20-30 days earlier than control without affecting yield, in glasshouse as well as net-house conditions, across seasons and generations. These changes in life cycle were associated with reciprocal changes in expression of GA pathway genes and basal GA levels between suppression and over-expression lines. SlERF36 interacted with the promoters of two GA2 oxidase genes, SlGA2ox3 and SlGA2ox4, and the DELLA gene, SlDELLA, reducing their transcription and causing a 3-5-fold increase in basal GA3/GA4 levels. Its suppression increased SlGA2ox3/4 transcript levels and reduced GA3/GA4 levels by 30%-50%. SlERF36 is conserved across families making it an important candidate in agricultural and horticultural crops for manipulation of plant growth and developmental transitions to reduce life cycles for faster harvest.
Keywords: AP2‐ERF domain; DELLA; GA2 oxidase; early maturity; flowering time; gibberellins.
© 2023 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no competing interests.
Figures
Similar articles
-
The EAR motif controls the early flowering and senescence phenotype mediated by over-expression of SlERF36 and is partly responsible for changes in stomatal density and photosynthesis.PLoS One. 2014 Jul 18;9(7):e101995. doi: 10.1371/journal.pone.0101995. eCollection 2014. PLoS One. 2014. PMID: 25036097 Free PMC article.
-
Tomato SlARF5 participate in the flower organ initiation process and control plant height.BMC Plant Biol. 2024 Oct 23;24(1):993. doi: 10.1186/s12870-024-05707-z. BMC Plant Biol. 2024. PMID: 39438786 Free PMC article.
-
Identification and functional study of a mild allele of SlDELLA gene conferring the potential for improved yield in tomato.Sci Rep. 2018 Aug 13;8(1):12043. doi: 10.1038/s41598-018-30502-w. Sci Rep. 2018. PMID: 30104574 Free PMC article.
-
Molecular, hormonal, and metabolic mechanisms of fruit set, the ovary-to-fruit transition, in horticultural crops.J Exp Bot. 2023 Oct 31;74(20):6254-6268. doi: 10.1093/jxb/erad214. J Exp Bot. 2023. PMID: 37279328 Review.
-
Molecular GA pathways as conserved integrators for adaptive responses.Plant Biol (Stuttg). 2023 Aug;25(5):649-660. doi: 10.1111/plb.13549. Epub 2023 Jun 15. Plant Biol (Stuttg). 2023. PMID: 37279043 Review.
Cited by
-
GA3-Induced SlXTH19 Expression Enhances Cell Wall Remodeling and Plant Height in Tomatoes.Plants (Basel). 2024 Dec 21;13(24):3578. doi: 10.3390/plants13243578. Plants (Basel). 2024. PMID: 39771276 Free PMC article.
-
Phylogeny and expression patterns of ERF genes that are potential reproductive inducers in hybrid larch.BMC Genomics. 2024 Mar 18;25(1):288. doi: 10.1186/s12864-024-10188-3. BMC Genomics. 2024. PMID: 38500084 Free PMC article.
-
Genome-Wide Analysis of CSL Family Genes Involved in Petiole Elongation, Floral Petalization, and Response to Salinity Stress in Nelumbo nucifera.Int J Mol Sci. 2024 Nov 22;25(23):12531. doi: 10.3390/ijms252312531. Int J Mol Sci. 2024. PMID: 39684243 Free PMC article.
-
Molecular breeding of tomato: Advances and challenges.J Integr Plant Biol. 2025 Mar;67(3):669-721. doi: 10.1111/jipb.13879. Epub 2025 Mar 18. J Integr Plant Biol. 2025. PMID: 40098531 Free PMC article. Review.
-
Genome-wide identification of SlIQMs and the regulatory effect of calcium on tomato seedlings under drought stress and phytohormone treatment.Plant Cell Rep. 2025 Mar 7;44(4):70. doi: 10.1007/s00299-025-03459-0. Plant Cell Rep. 2025. PMID: 40055201
References
-
- Abel, S. and Theologis, A. (1994) Transient transformation of Arabidopsis leaf protoplasts: a versatile experimental system to study gene expression. Plant J. 5, 421–427. - PubMed
-
- Achard, P. , Cheng, H. , De Grauwe, L. , Decat, J. , Schoutteten, H. , Moritz, T. , Van Der Straeten, D. et al. (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94. - PubMed
-
- Asif, M.H. , Dhawan, P. and Nath, P. (2000) A simple procedure for the isolation of high quality RNA from ripening banana fruit. Plant Mol. Biol. Rep. 18, 109–115.
-
- Bauerle, W.L. (2022) Gibberellin A3 induced flowering intensification in Humulus lupulus L.: Synchronizing vegetative phase change and photoperiod induction. Sci. Hortic., 302, 111183.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

