Unique adipose tissue invariant natural killer T cell subpopulations control adipocyte turnover in mice
- PMID: 38129377
- PMCID: PMC10739728
- DOI: 10.1038/s41467-023-44181-3
Unique adipose tissue invariant natural killer T cell subpopulations control adipocyte turnover in mice
Abstract
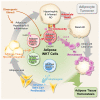
Adipose tissue invariant natural killer T (iNKT) cells are a crucial cell type for adipose tissue homeostasis in obese animals. However, heterogeneity of adipose iNKT cells and their function in adipocyte turnover are not thoroughly understood. Here, we investigate transcriptional heterogeneity in adipose iNKT cells and their hierarchy using single-cell RNA sequencing in lean and obese mice. We report that distinct subpopulations of adipose iNKT cells modulate adipose tissue homeostasis through adipocyte death and birth. We identify KLRG1+ iNKT cells as a unique iNKT cell subpopulation in adipose tissue. Adoptive transfer experiments showed that KLRG1+ iNKT cells are selectively generated within adipose tissue microenvironment and differentiate into a CX3CR1+ cytotoxic subpopulation in obese mice. In addition, CX3CR1+ iNKT cells specifically kill enlarged and inflamed adipocytes and recruit macrophages through CCL5. Furthermore, adipose iNKT17 cells have the potential to secrete AREG, and AREG is involved in stimulating adipose stem cell proliferation. Collectively, our data suggest that each adipose iNKT cell subpopulation plays key roles in the control of adipocyte turnover via interaction with adipocytes, adipose stem cells, and macrophages in adipose tissue.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

