Circulating small extracellular vesicles microRNAs plus CA-125 for treatment stratification in advanced ovarian cancer
- PMID: 38129848
- PMCID: PMC10740240
- DOI: 10.1186/s12967-023-04774-4
Circulating small extracellular vesicles microRNAs plus CA-125 for treatment stratification in advanced ovarian cancer
Abstract
Background: No residual disease (R0 resection) after debulking surgery is the most critical independent prognostic factor for advanced ovarian cancer (AOC). There is an unmet clinical need for selecting primary or interval debulking surgery in AOC patients using existing prediction models.
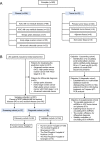
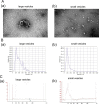
Methods: RNA sequencing of circulating small extracellular vesicles (sEVs) was used to discover the differential expression microRNAs (DEMs) profile between any residual disease (R0, n = 17) and no residual disease (non-R0, n = 20) in AOC patients. We further analyzed plasma samples of AOC patients collected before surgery or neoadjuvant chemotherapy via TaqMan qRT-PCR. The combined risk model of residual disease was developed by logistic regression analysis based on the discovery-validation sets.
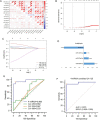
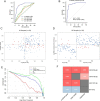
Results: Using a comprehensive plasma small extracellular vesicles (sEVs) microRNAs (miRNAs) profile in AOC, we identified and optimized a risk prediction model consisting of plasma sEVs-derived 4-miRNA and CA-125 with better performance in predicting R0 resection. Based on 360 clinical human samples, this model was constructed using least absolute shrinkage and selection operator (LASSO) and logistic regression analysis, and it has favorable calibration and discrimination ability (AUC:0.903; sensitivity:0.897; specificity:0.910; PPV:0.926; NPV:0.871). The quantitative evaluation of Net Reclassification Improvement (NRI) and Integrated Discrimination Improvement (IDI) suggested that the additional predictive power of the combined model was significantly improved contrasted with CA-125 or 4-miRNA alone (NRI = 0.471, IDI = 0.538, p < 0.001; NRI = 0.122, IDI = 0.185, p < 0.01).
Conclusion: Overall, we established a reliable, non-invasive, and objective detection method composed of circulating tumor-derived sEVs 4-miRNA plus CA-125 to preoperatively anticipate the high-risk AOC patients of residual disease to optimize clinical therapy.
Keywords: Ovarian cancer; Prediction model; Residual disease; Small extracellular vesicles; microRNA.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Horowitz NS, Miller A, Rungruang B, Richard SD, Rodriguez N, Bookman MA, Hamilton CA, Krivak TC, Maxwell GL. Does aggressive surgery improve outcomes? Interaction between preoperative disease burden and complex surgery in patients with advanced-stage ovarian cancer: an analysis of gog 182. J Clin Oncol. 2015;33:937–943. doi: 10.1200/JCO.2014.56.3106. - DOI - PMC - PubMed
-
- William EWI, Maxwell GL, Tian C, Sundborg MJ, Rose GS, Rose PG, Rubin SC, Muggia F, Mcguire WP, Gynecologic OG. Tumor residual after surgical cytoreduction in prediction of clinical outcome in stage iv epithelial ovarian cancer: a gynecologic oncology group study. J Clin Oncol. 2008;26:83–89. doi: 10.1200/JCO.2007.13.1953. - DOI - PubMed
-
- Vergote I, Coens C, Nankivell M, Kristensen GB, Parmar M, Ehlen T, Jayson GC, Johnson N, Swart AM, Verheijen R, Mccluggage WG, Perren T, Panici PB, Kenter G, Casado A, Mendiola C, Stuart G, Reed NS, Kehoe S. Neoadjuvant chemotherapy versus debulking surgery in advanced tubo-ovarian cancers: pooled analysis of individual patient data from the eortc 55971 and chorus trials. Lancet Oncol. 2018;19:1680–1687. doi: 10.1016/S1470-2045(18)30566-7. - DOI - PubMed
-
- Suidan RS, Ramirez PT, Sarasohn DM, Teitcher JB, Iyer RB, Zhou Q, Iasonos A, Denesopolis J, Zivanovic O, Long Roche KC, Sonoda Y, Coleman RL, Abu-Rustum NR, Hricak H, Chi DS. A multicenter assessment of the ability of preoperative computed tomography scan and ca-125 to predict gross residual disease at primary debulking for advanced epithelial ovarian cancer. Gynecol Oncol. 2017;145:27–31. doi: 10.1016/j.ygyno.2017.02.020. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- No. 2020JJ4051/the General Project of Natural Science Foundation of Hunan Province
- No. WZ2020-15/Promotion Project of Health Suitability Program in Health Department of Hunan Province
- No. 2020SK51101/Science and Technology Innovation Program of Hunan Province
- No. ZX2020004/Hunan Cancer Hospital Climbing Fund
- No. 20201127-1001/Capacity Building Project of Central Subsidy Medical and Health Institutions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

