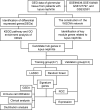
Identification of driver genes in lupus nephritis based on comprehensive bioinformatics and machine learning
- PMID: 38130724
- PMCID: PMC10733527
- DOI: 10.3389/fimmu.2023.1288699
Identification of driver genes in lupus nephritis based on comprehensive bioinformatics and machine learning
Abstract
Background: Lupus nephritis (LN) is a common and severe glomerulonephritis that often occurs as an organ manifestation of systemic lupus erythematosus (SLE). However, the complex pathological mechanisms associated with LN have hindered the progress of targeted therapies.
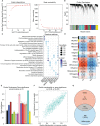
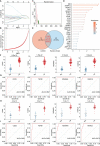
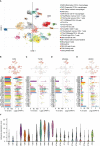
Methods: We analyzed glomerular tissues from 133 patients with LN and 51 normal controls using data obtained from the GEO database. Differentially expressed genes (DEGs) were identified and subjected to enrichment analysis. Weighted gene co-expression network analysis (WGCNA) was utilized to identify key gene modules. The least absolute shrinkage and selection operator (LASSO) and random forest were used to identify hub genes. We also analyzed immune cell infiltration using CIBERSORT. Additionally, we investigated the relationships between hub genes and clinicopathological features, as well as examined the distribution and expression of hub genes in the kidney.
Results: A total of 270 DEGs were identified in LN. Using weighted gene co-expression network analysis (WGCNA), we clustered these DEGs into 14 modules. Among them, the turquoise module displayed a significant correlation with LN (cor=0.88, p<0.0001). Machine learning techniques identified four hub genes, namely CD53 (AUC=0.995), TGFBI (AUC=0.997), MS4A6A (AUC=0.994), and HERC6 (AUC=0.999), which are involved in inflammation response and immune activation. CIBERSORT analysis suggested that these hub genes may contribute to immune cell infiltration. Furthermore, these hub genes exhibited strong correlations with the classification, renal function, and proteinuria of LN. Interestingly, the highest hub gene expression score was observed in macrophages.
Conclusion: CD53, TGFBI, MS4A6A, and HERC6 have emerged as promising candidate driver genes for LN. These hub genes hold the potential to offer valuable insights into the molecular diagnosis and treatment of LN.
Keywords: Lupus nephritis; WGCNA; bioinformatics; immune infiltration; machine learning.
Copyright © 2023 Wang, Hu, Pei, Zeng and Yao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Predicting diagnostic gene expression profiles associated with immune infiltration in patients with lupus nephritis.Front Immunol. 2022 Dec 2;13:839197. doi: 10.3389/fimmu.2022.839197. eCollection 2022. Front Immunol. 2022. PMID: 36532018 Free PMC article.
-
Identification and validation of key autophagy-related genes in lupus nephritis by bioinformatics and machine learning.PLoS One. 2025 Jan 27;20(1):e0318280. doi: 10.1371/journal.pone.0318280. eCollection 2025. PLoS One. 2025. PMID: 39869603 Free PMC article.
-
Machine learning-based identification of novel hub genes associated with oxidative stress in lupus nephritis: implications for diagnosis and therapeutic targets.Lupus Sci Med. 2024 Apr 18;11(1):e001126. doi: 10.1136/lupus-2023-001126. Lupus Sci Med. 2024. PMID: 38637124 Free PMC article.
-
Transcription Factors in the Pathogenesis of Lupus Nephritis and Their Targeted Therapy.Int J Mol Sci. 2024 Jan 16;25(2):1084. doi: 10.3390/ijms25021084. Int J Mol Sci. 2024. PMID: 38256157 Free PMC article. Review.
-
Role of Traditional Chinese Medicine for the Treatment of Lupus nephritis: Mechanisms and Applications.Altern Ther Health Med. 2024 Jun;30(6):154-165. Altern Ther Health Med. 2024. PMID: 37944951 Review.
Cited by
-
Lupus Nephritis from Pathogenesis to New Therapies: An Update.Int J Mol Sci. 2024 Aug 18;25(16):8981. doi: 10.3390/ijms25168981. Int J Mol Sci. 2024. PMID: 39201667 Free PMC article. Review.
-
Identification of ALDH2 as a novel target for the treatment of acute kidney injury in kidney transplantation based on WGCNA and machine learning algorithms and exploration of its potential mechanism of action using animal experiments.Front Immunol. 2025 Mar 4;16:1536800. doi: 10.3389/fimmu.2025.1536800. eCollection 2025. Front Immunol. 2025. PMID: 40103812 Free PMC article.
-
Lupus Nephritis Biomarkers: A Critical Review.Int J Mol Sci. 2024 Jan 9;25(2):805. doi: 10.3390/ijms25020805. Int J Mol Sci. 2024. PMID: 38255879 Free PMC article. Review.
-
TGF-β/JNK axis mediates mitochondrial damage and macrophage cGAS-STING activation in liver Mallory-Denk body pathogenesis.J Transl Med. 2025 Jun 3;23(1):621. doi: 10.1186/s12967-025-06618-9. J Transl Med. 2025. PMID: 40462149 Free PMC article.
-
A potential XGBoost Diagnostic Score for Staphylococcus aureus bloodstream infection.Front Immunol. 2025 Apr 22;16:1574003. doi: 10.3389/fimmu.2025.1574003. eCollection 2025. Front Immunol. 2025. PMID: 40330459 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

