Separable Roles for Neur and Ubiquitin in Delta Signalling in the Drosophila CNS Lineages
- PMID: 38132160
- PMCID: PMC10741450
- DOI: 10.3390/cells12242833
Separable Roles for Neur and Ubiquitin in Delta Signalling in the Drosophila CNS Lineages
Abstract
The execution of a Notch signal at the plasma membrane relies on the mechanical force exerted onto Notch by its ligand. It has been appreciated that the DSL ligands need to collaborate with a ubiquitin (Ub) ligase, either Neuralized or Mindbomb1, in order to exert this pulling force, but the role of ubiquitylation per se is uncertain. Regarding the Delta-Neur pair, it is documented that neither the Neur catalytic domain nor the Delta intracellular lysines (putative Ub acceptors) are needed for activity. Here, we present a dissection of the Delta activity using the Delta-Notch-dependent expression of Hey in newborn Drosophila neurons as a sensitive in vivo assay. We show that the Delta-Neur interaction per se, rather than ubiquitylation, is needed for activity, pointing to the existence of a Delta-Neur signaling complex. The Neur catalytic domain, although not strictly needed, greatly improves Delta-Neur complex functionality when the Delta lysines are mutated, suggesting that the ubiquitylation of some component of the complex, other than Delta, can enhance signaling. Since Hey expression is sensitive to the perturbation of endocytosis, we propose that the Delta-Neur complex triggers a force-generating endocytosis event that activates Notch in the adjacent cell.
Keywords: Delta; Drosophila; Hey; Neuralized; Notch; asymmetric cell division; endocytosis; neurodevelopment; ubiquitylation.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

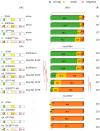

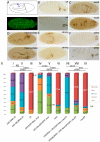
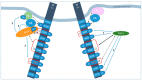
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

