Antiviral Activity of Zinc Finger Antiviral Protein (ZAP) in Different Virus Families
- PMID: 38133344
- PMCID: PMC10747524
- DOI: 10.3390/pathogens12121461
Antiviral Activity of Zinc Finger Antiviral Protein (ZAP) in Different Virus Families
Abstract
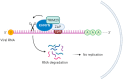
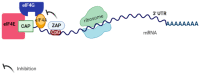
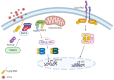
The CCCH-type zinc finger antiviral protein (ZAP) in humans, specifically isoforms ZAP-L and ZAP-S, is a crucial component of the cell's intrinsic immune response. ZAP acts as a post-transcriptional RNA restriction factor, exhibiting its activity during infections caused by retroviruses and alphaviruses. Its function involves binding to CpG (cytosine-phosphate-guanine) dinucleotide sequences present in viral RNA, thereby directing it towards degradation. Since vertebrate cells have a suppressed frequency of CpG dinucleotides, ZAP is capable of distinguishing foreign genetic elements. The expression of ZAP leads to the reduction of viral replication and impedes the assembly of new virus particles. However, the specific mechanisms underlying these effects have yet to be fully understood. Several questions regarding ZAP's mechanism of action remain unanswered, including the impact of CpG dinucleotide quantity on ZAP's activity, whether this sequence is solely required for the binding between ZAP and viral RNA, and whether the recruitment of cofactors is dependent on cell type, among others. This review aims to integrate the findings from studies that elucidate ZAP's antiviral role in various viral infections, discuss gaps that need to be filled through further studies, and shed light on new potential targets for therapeutic intervention.
Keywords: PARP-13; ZAP protein; antiviral activity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Versatility of the Zinc-Finger Antiviral Protein (ZAP) As a Modulator of Viral Infections.Int J Biol Sci. 2024 Aug 26;20(12):4585-4600. doi: 10.7150/ijbs.98029. eCollection 2024. Int J Biol Sci. 2024. PMID: 39309436 Free PMC article. Review.
-
Association of Zinc Finger Antiviral Protein Binding to Viral Genomic RNA with Attenuation of Replication of Echovirus 7.mSphere. 2021 Jan 6;6(1):e01138-20. doi: 10.1128/mSphere.01138-20. mSphere. 2021. PMID: 33408233 Free PMC article.
-
Riplet Binds the Zinc Finger Antiviral Protein (ZAP) and Augments ZAP-Mediated Restriction of HIV-1.J Virol. 2022 Aug 24;96(16):e0052622. doi: 10.1128/jvi.00526-22. Epub 2022 Aug 1. J Virol. 2022. PMID: 35913217 Free PMC article.
-
ZAP's stress granule localization is correlated with its antiviral activity and induced by virus replication.PLoS Pathog. 2019 May 22;15(5):e1007798. doi: 10.1371/journal.ppat.1007798. eCollection 2019 May. PLoS Pathog. 2019. PMID: 31116799 Free PMC article.
-
Does the Zinc Finger Antiviral Protein (ZAP) Shape the Evolution of Herpesvirus Genomes?Viruses. 2021 Sep 17;13(9):1857. doi: 10.3390/v13091857. Viruses. 2021. PMID: 34578438 Free PMC article. Review.
Cited by
-
Evolutionary analysis of ZAP and its cofactors identifies intrinsically disordered regions as central elements in host-pathogen interactions.Comput Struct Biotechnol J. 2024 Aug 2;23:3143-3154. doi: 10.1016/j.csbj.2024.07.022. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 39234301 Free PMC article.
-
Host RNA-Binding Proteins as Regulators of HIV-1 Replication.Viruses. 2024 Dec 31;17(1):43. doi: 10.3390/v17010043. Viruses. 2024. PMID: 39861832 Free PMC article. Review.
-
Impact of a Functional Dairy Powder and Its Primary Component on the Growth of Pathogenic and Probiotic Gut Bacteria and Human Coronavirus 229E.Int J Mol Sci. 2024 Aug 29;25(17):9353. doi: 10.3390/ijms25179353. Int J Mol Sci. 2024. PMID: 39273301 Free PMC article.
-
Impact of the Zinc Antiviral Protein on the Genomic Composition of RNA Viruses Infecting Vertebrates.Mol Biol Evol. 2025 Jun 4;42(6):msaf135. doi: 10.1093/molbev/msaf135. Mol Biol Evol. 2025. PMID: 40465846 Free PMC article.
-
Dual Roles of Host Zinc Finger Proteins in Viral RNA Regulation: Decay or Stabilization.Int J Mol Sci. 2024 Oct 17;25(20):11138. doi: 10.3390/ijms252011138. Int J Mol Sci. 2024. PMID: 39456919 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources

