Predicting disease severity in multiple sclerosis using multimodal data and machine learning
- PMID: 38133801
- PMCID: PMC10896787
- DOI: 10.1007/s00415-023-12132-z
Predicting disease severity in multiple sclerosis using multimodal data and machine learning
Abstract
Background: Multiple sclerosis patients would benefit from machine learning algorithms that integrates clinical, imaging and multimodal biomarkers to define the risk of disease activity.
Methods: We have analysed a prospective multi-centric cohort of 322 MS patients and 98 healthy controls from four MS centres, collecting disability scales at baseline and 2 years later. Imaging data included brain MRI and optical coherence tomography, and omics included genotyping, cytomics and phosphoproteomic data from peripheral blood mononuclear cells. Predictors of clinical outcomes were searched using Random Forest algorithms. Assessment of the algorithm performance was conducted in an independent prospective cohort of 271 MS patients from a single centre.
Results: We found algorithms for predicting confirmed disability accumulation for the different scales, no evidence of disease activity (NEDA), onset of immunotherapy and the escalation from low- to high-efficacy therapy with intermediate to high-accuracy. This accuracy was achieved for most of the predictors using clinical data alone or in combination with imaging data. Still, in some cases, the addition of omics data slightly increased algorithm performance. Accuracies were comparable in both cohorts.
Conclusion: Combining clinical, imaging and omics data with machine learning helps identify MS patients at risk of disability worsening.
Keywords: Imaging; Machine learning; Multiple sclerosis; Omics; Precision medicine.
© 2023. The Author(s).
Conflict of interest statement
Magi Andorra is an employee of Hoffman-La Roche AG. Yet this article is related to his activity at the Hospital Clinic of Barcelona. Ana Freire reports no disclosures. Irati Zubizarreta received reimbursement from Genzyme, Biogen, Merck, and Bayer-Schering. Irene Pulido-Valdeolivas is currently an employee of UCB pharma. Yet this article is related to her activity at the Hospital Clinic of Barcelona. She has received travel reimbursement from Roche Spain and Genzyme-Sanofi, European Academy of Neurology, and European Committee for Treatment and Research in Multiple Sclerosis for international and national meetings over the last 3 years; she holds a patent for an affordable eye-tracking system to measure eye movement in neurologic diseases, and she holds stock in Aura Innovative Robotics. Elena H Martinez-Lapiscina is an employee of the European Medicines Agency (Human Medicines) since 16 April 2019. Yet this article is related to her activity at the Hospital Clinic of Barcelona and consequently. It does not in any way represent the views of the Agency or its Committees. Sara Llufriu received compensation for consulting services and speaker honoraria from Biogen Idec, Novartis, TEVA, Genzyme, Sanofi, and Merck. Albert Saiz received compensation for consulting services and speaker honoraria from Bayer-Schering, Merck-Serono, Biogen-Idec, Sanofi-Aventis, TEVA, Novartis, and Roche. Eloy Martinez-Heras reports no disclosures. Elisabeth Solana received travel reimbursement from Sanofi and ECTRIMS and reports personal fees from Roche Spain. Melanie Rinas reports no disclosures. Julio Saez-Rodriguez reports no disclosures. Steffan Bos reports no disclosures. Maria Cellerino reports no disclosures. Federico Ivaldi reports no disclosures. Matteo Pardini received research support from Novartis and Nutricia and honoraria from Merk and Novartis. Gemma Vila reports no disclosures. Sigrid A. de Rodez Benavent reports no disclosures. Synne Brune Ingebetsen has received honoraria for lecturing from Biogen and Novartis. Priscilla Bäcker-Koduah is funded by the DFG Excellence grant to FP (DFG exc 257) and is a Junior scholar of the Einstein Foundation. Tone Berge has received unrestricted research grants from Biogen and Sanofi-Genzyme. Einar Høgestøl received honoraria for lecturing and advisory board activity from Biogen, MS-union, Merck, and Sanofi-Genzyme and unrestricted research grant from Merck. Friedemann Paul received honoraria and research support from Alexion, Bayer, Biogen, Chugai, Merck Serono, Novartis, Genzyme, MedImmune, Shire, Teva, and serves on scientific advisory boards for Alexion, MedImmune, and Novartis. He has received funding from Deutsche Forschungsgemeinschaft (DFG Exc 257), Bundesministerium für Bildung und Forschung (Competence Network Multiple Sclerosis), Guthy Jackson Charitable Foundation, EU Framework Program 7, National Multiple Sclerosis Society of the USA. Hanne F. Harbo reports no disclosures. Nicole Kerlero de Rosbo reports no disclosures. Claudia Chien received honoraria for speaking from Bayer and research funding from Novartis, unrelated to this study. Susanna Asseyer received a conference grant from Celgene and honoraria for speaking from Alexion, Bayer, and Roche. Janina Behrens reports no disclosures. Alex Brandt has a patent pending for Perceptive visual computing-based postural control analysis, Multiple sclerosis biomarker, Perceptive sleep motion analysis, and Fovea morphometry; consulted for Motognosis; is on the executive board of IMSVISUAL; received research support from Novartis, Biogen, BMWi, BMBF, and the Guthy-Jackson Charitable Foundation; and holds stock or stock options in Motognosis. Leonidas G Alexopoulos is founder and hold stocks at ProtATonce. Antonio Uccelli received grants and contracts from FISM, Novartis, Biogen, Merck, Fondazione Cariplo, Italian Ministry of Health, received honoraria, or consultation fees from Biogen, Roche, Teva, Merck, Genzyme, Novartis. Ricardo Baeza-Yates reports no disclosures. Pablo Villoslada has received consultancy fees and hold stocks from Accure Therapeutics SL, Attune Neurosciences Inc, Spiral Therapeutics Inc, QMenta Inc, CLight Inc, NeuroPrex Inc, Oculis SA and Adhera Health Inc. Other authors do not have competing interests.
Figures

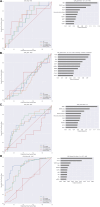
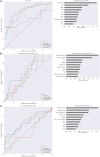
References
-
- Pulido-Valdeolivas I, Zubizarreta I, Martinez-Lapiscina E, Villoslada P. Precision medicine for multiple sclerosis: an update of the available biomarkers and their use in therapeutic decision making. Expert Rev Precis Med Drug Dev. 2017;2:1–17. doi: 10.1080/23808993.2017.1393315. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

