Chromosome 8p engineering reveals increased metastatic potential targetable by patient-specific synthetic lethality in liver cancer
- PMID: 38134284
- PMCID: PMC10745716
- DOI: 10.1126/sciadv.adh1442
Chromosome 8p engineering reveals increased metastatic potential targetable by patient-specific synthetic lethality in liver cancer
Abstract
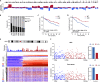
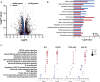
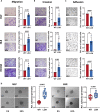
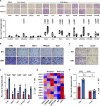
Large-scale chromosomal aberrations are prevalent in human cancer, but their function remains poorly understood. We established chromosome-engineered hepatocellular carcinoma cell lines using CRISPR-Cas9 genome editing. A 33-mega-base pair region on chromosome 8p (chr8p) was heterozygously deleted, mimicking a frequently observed chromosomal deletion. Using this isogenic model system, we delineated the functional consequences of chr8p loss and its impact on metastatic behavior and patient survival. We found that metastasis-associated genes on chr8p act in concert to induce an aggressive and invasive phenotype characteristic for chr8p-deleted tumors. Genome-wide CRISPR-Cas9 viability screening in isogenic chr8p-deleted cells served as a powerful tool to find previously unidentified synthetic lethal targets and vulnerabilities accompanying patient-specific chromosomal alterations. Using this target identification strategy, we showed that chr8p deletion sensitizes tumor cells to targeting of the reactive oxygen sanitizing enzyme Nudix hydrolase 17. Thus, chromosomal engineering allowed for the identification of novel synthetic lethalities specific to chr8p loss of heterozygosity.
Figures




References
-
- D. Hanahan, Hallmarks of cancer: New dimensions. Cancer Discov. 12, 31–46 (2022). - PubMed
-
- A. M. Taylor, J. Shih, G. Ha, G. F. Gao, X. Zhang, A. C. Berger, S. E. Schumacher, C. Wang, H. Hu, J. Liu, A. J. Lazar; The Cancer Genome Atlas Research Network, A. D. Cherniack, R. Beroukhim, M. Meyerson, Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell 33, 676–689 e3 (2018). - PMC - PubMed
-
- R. Beroukhim, C. H. Mermel, D. Porter, G. Wei, S. Raychaudhuri, J. Donovan, J. Barretina, J. S. Boehm, J. Dobson, M. Urashima, K. T. Mc Henry, R. M. Pinchback, A. H. Ligon, Y. J. Cho, L. Haery, H. Greulich, M. Reich, W. Winckler, M. S. Lawrence, B. A. Weir, K. E. Tanaka, D. Y. Chiang, A. J. Bass, A. Loo, C. Hoffman, J. Prensner, T. Liefeld, Q. Gao, D. Yecies, S. Signoretti, E. Maher, F. J. Kaye, H. Sasaki, J. E. Tepper, J. A. Fletcher, J. Tabernero, J. Baselga, M. S. Tsao, F. Demichelis, M. A. Rubin, P. A. Janne, M. J. Daly, C. Nucera, R. L. Levine, B. L. Ebert, S. Gabriel, A. K. Rustgi, C. R. Antonescu, M. Ladanyi, A. Letai, L. A. Garraway, M. Loda, D. G. Beer, L. D. True, A. Okamoto, S. L. Pomeroy, S. Singer, T. R. Golub, E. S. Lander, G. Getz, W. R. Sellers, M. Meyerson, The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010). - PMC - PubMed
-
- K. L. Gorringe, M. Ramakrishna, L. H. Williams, A. Sridhar, S. E. Boyle, J. L. Bearfoot, J. Li, M. S. Anglesio, I. G. Campbell, Are there any more ovarian tumor suppressor genes? A new perspective using ultra high-resolution copy number and loss of heterozygosity analysis. Genes Chromosomes Cancer 48, 931–942 (2009). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

