Delayed Mucosal Antiviral Responses Despite Robust Peripheral Inflammation in Fatal COVID-19
- PMID: 39052740
- PMCID: PMC11272059
- DOI: 10.1093/infdis/jiad590
Delayed Mucosal Antiviral Responses Despite Robust Peripheral Inflammation in Fatal COVID-19
Abstract
Background: While inflammatory and immune responses to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in peripheral blood are extensively described, responses at the upper respiratory mucosal site of initial infection are relatively poorly defined. We sought to identify mucosal cytokine/chemokine signatures that distinguished coronavirus disease 2019 (COVID-19) severity categories, and relate these to disease progression and peripheral inflammation.
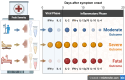
Methods: We measured 35 cytokines and chemokines in nasal samples from 274 patients hospitalized with COVID-19. Analysis considered the timing of sampling during disease, as either the early (0-5 days after symptom onset) or late (6-20 days after symptom onset) phase.
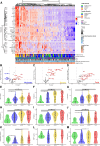
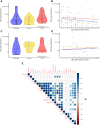
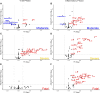
Results: Patients that survived severe COVID-19 showed interferon (IFN)-dominated mucosal immune responses (IFN-γ, CXCL10, and CXCL13) early in infection. These early mucosal responses were absent in patients who would progress to fatal disease despite equivalent SARS-CoV-2 viral load. Mucosal inflammation in later disease was dominated by interleukin 2 (IL-2), IL-10, IFN-γ, and IL-12p70, which scaled with severity but did not differentiate patients who would survive or succumb to disease. Cytokines and chemokines in the mucosa showed distinctions from responses evident in the peripheral blood, particularly during fatal disease.
Conclusions: Defective early mucosal antiviral responses anticipate fatal COVID-19 but are not associated with viral load. Early mucosal immune responses may define the trajectory of severe COVID-19.
Keywords: COVID-19; SARS-CoV-2; airway; chemokine; cytokine; lung; mucosa; nose; virus.
© The Author(s) 2023. Published by Oxford University Press on behalf of Infectious Diseases Society of America.
Conflict of interest statement
Potential conflicts of interest. P. J. M. O. is the Imperial College Lead Investigator of the EMINENT consortium, supported by the Medical Research Council UK (MR/R502121/1; this grant supports collaborative testing of compounds developed by GlaxoSmithKline in UK universities); is on advisory boards for Affnivax, Oxford Immunotech, Nestle, and Pfizer in relation to immunity to viruses (fees paid to Imperial College London); and is on an advisory board for Janssen/J&J. L.T. has received consulting fees from the Medicines and Healthcare products Regulatory Agency (MHRA); consulting fees from AstraZeneca and Synairgen, paid to the University of Liverpool; speaker’s fees from Eisai, Ltd; and support for conference attendance from AstraZeneca. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

