Epithelial SIRT6 governs IL-17A pathogenicity and drives allergic airway inflammation and remodeling
- PMID: 38135684
- PMCID: PMC10746710
- DOI: 10.1038/s41467-023-44179-x
Epithelial SIRT6 governs IL-17A pathogenicity and drives allergic airway inflammation and remodeling
Abstract
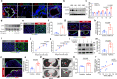
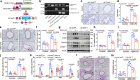
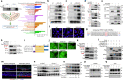
Dysregulation of IL-17A is closely associated with airway inflammation and remodeling in severe asthma. However, the molecular mechanisms by which IL-17A is regulated remain unclear. Here we identify epithelial sirtuin 6 (SIRT6) as an epigenetic regulator that governs IL-17A pathogenicity in severe asthma. Mice with airway epithelial cell-specific deletion of Sirt6 are protected against allergen-induced airway inflammation and remodeling via inhibiting IL-17A-mediated inflammatory chemokines and mesenchymal reprogramming. Mechanistically, SIRT6 directly interacts with RORγt and mediates RORγt deacetylation at lysine 192 via its PPXY motifs. SIRT6 promotes RORγt recruitment to the IL-17A gene promoter and enhances its transcription. In severe asthma patients, high expression of SIRT6 positively correlates with airway remodeling and disease severity. SIRT6 inhibitor (OSS_128167) treatment significantly attenuates airway inflammation and remodeling in mice. Collectively, these results uncover a function for SIRT6 in regulating IL-17A pathogenicity in severe asthma, implicating SIRT6 as a potential therapeutic target for severe asthma.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

