YidC from Escherichia coli Forms an Ion-Conducting Pore upon Activation by Ribosomes
- PMID: 38136645
- PMCID: PMC10741985
- DOI: 10.3390/biom13121774
YidC from Escherichia coli Forms an Ion-Conducting Pore upon Activation by Ribosomes
Abstract
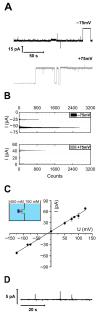
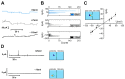
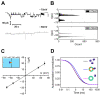
The universally conserved protein YidC aids in the insertion and folding of transmembrane polypeptides. Supposedly, a charged arginine faces its hydrophobic lipid core, facilitating polypeptide sliding along YidC's surface. How the membrane barrier to other molecules may be maintained is unclear. Here, we show that the purified and reconstituted E. coli YidC forms an ion-conducting transmembrane pore upon ribosome or ribosome-nascent chain complex (RNC) binding. In contrast to monomeric YidC structures, an AlphaFold parallel YidC dimer model harbors a pore. Experimental evidence for a dimeric assembly comes from our BN-PAGE analysis of native vesicles, fluorescence correlation spectroscopy studies, single-molecule fluorescence photobleaching observations, and crosslinking experiments. In the dimeric model, the conserved arginine and other residues interacting with nascent chains point into the putative pore. This result suggests the possibility of a YidC-assisted insertion mode alternative to the insertase mechanism.
Keywords: electrophysiology; fluorescence correlation spectroscopy; protein translocation; single dye tracing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
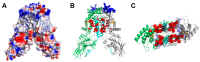

Similar articles
-
Structural Dynamics of the YidC:Ribosome Complex during Membrane Protein Biogenesis.Cell Rep. 2016 Dec 13;17(11):2943-2954. doi: 10.1016/j.celrep.2016.11.059. Cell Rep. 2016. PMID: 27974208 Free PMC article.
-
Elucidating the native architecture of the YidC: ribosome complex.J Mol Biol. 2013 Nov 15;425(22):4112-24. doi: 10.1016/j.jmb.2013.07.042. Epub 2013 Aug 8. J Mol Biol. 2013. PMID: 23933010
-
A structural model of the active ribosome-bound membrane protein insertase YidC.Elife. 2014 Jul 10;3:e03035. doi: 10.7554/eLife.03035. Elife. 2014. PMID: 25012291 Free PMC article.
-
The ribosome and YidC. New insights into the biogenesis of Escherichia coli inner membrane proteins.EMBO Rep. 2003 Oct;4(10):939-43. doi: 10.1038/sj.embor.embor921. EMBO Rep. 2003. PMID: 14528263 Free PMC article. Review.
-
The versatile role of YidC in membrane protein biosynthesis and quality control.Biochim Biophys Acta Mol Cell Res. 2025 Jun;1872(5):119956. doi: 10.1016/j.bbamcr.2025.119956. Epub 2025 Apr 10. Biochim Biophys Acta Mol Cell Res. 2025. PMID: 40221051 Review.
Cited by
-
Deciphering the Interdomain Coupling in a Gram-Negative Bacterial Membrane Insertase.J Phys Chem B. 2024 Oct 10;128(40):9734-9744. doi: 10.1021/acs.jpcb.4c02824. Epub 2024 Sep 27. J Phys Chem B. 2024. PMID: 39329451 Free PMC article.
-
Enhanced Site-Specific Fluorescent Labeling of Membrane Proteins Using Native Nanodiscs.Biomolecules. 2025 Feb 10;15(2):254. doi: 10.3390/biom15020254. Biomolecules. 2025. PMID: 40001557 Free PMC article.
-
Cardiolipin occupancy profiles of YidC paralogs reveal the significance of respective TM2 helix residues in determining paralog-specific phenotypes.Front Mol Biosci. 2023 Oct 6;10:1264454. doi: 10.3389/fmolb.2023.1264454. eCollection 2023. Front Mol Biosci. 2023. PMID: 37867558 Free PMC article.
References
-
- Samuelson J.C., Jiang F., Yi L., Chen M., de Gier J.W., Kuhn A., Dalbey R.E. Function of YidC for the insertion of M13 procoat protein in Escherichia coli: Translocation of mutants that show differences in their membrane potential dependence and Sec requirement. J. Biol. Chem. 2001;276:34847–34852. doi: 10.1074/jbc.M105793200. - DOI - PubMed
-
- Welte T., Kudva R., Kuhn P., Sturm L., Braig D., Muller M., Warscheid B., Drepper F., Koch H.G. Promiscuous targeting of polytopic membrane proteins to SecYEG or YidC by the Escherichia coli signal recognition particle. Mol. Biol. Cell. 2012;23:464–479. doi: 10.1091/mbc.e11-07-0590. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

