Identification of Potential miRNA-mRNA Regulatory Network Associated with Growth and Development of Hair Follicles in Forest Musk Deer
- PMID: 38136906
- PMCID: PMC10740511
- DOI: 10.3390/ani13243869
Identification of Potential miRNA-mRNA Regulatory Network Associated with Growth and Development of Hair Follicles in Forest Musk Deer
Abstract
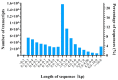
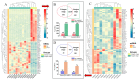
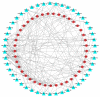
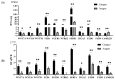
In this study, sRNA libraries and mRNA libraries of HFs of FMD were constructed and sequenced using an Illumina HiSeq 2500, and the expression profiles of miRNAs and genes in the HFs of FMD were obtained at the anagen and catagen stages. In total, 565 differentially expressed unigenes (DEGs) were identified, 90 of which were upregulated and 475 of which were downregulated. In the BP category of GO enrichment, the DEGs were enriched in the processes related to HF development and differentiation, including the hair cycle regulation and processes, HF development, skin epidermis development, regulation of HF development, skin development, the Wnt signaling pathway, and the BMP signaling pathway. Through KEGG analysis it was found that DEGs were significantly enriched in pathways associated with HF development and growth. A total of 186 differentially expressed miRNAs (DEmiRNAs) were screened (p < 0.05) in the HFs of FMD at the anagen stage vs. the catagen stage, 33 of which were upregulated and 153 of which were downregulated. Through DEmiRNA-mRNA association analysis, we found DEmiRNAs and target genes that mainly play regulatory roles in HF development and growth. The enrichment analysis of DEmiRNA target genes revealed similarities with the enrichment results of DEGs associated with HF development. Notably, both sets of genes were enriched in key pathways such as the Notch signaling pathway, melanogenesis, the cAMP signaling pathway, and cGMP-PKG. To validate our findings, we selected 11 DEGs and 11 DEmiRNAs for experimental verification using RT-qPCR. The results of the experimental validation were consistent with the RNA-Seq results.
Keywords: RT-qPCR; forest musk deer; hair follicle; miRNA–mRNA network; signal pathway analysis; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Integrated mRNA-miRNA transcriptome profiling of blood immune responses potentially related to pulmonary fibrosis in forest musk deer.Front Immunol. 2024 May 30;15:1404108. doi: 10.3389/fimmu.2024.1404108. eCollection 2024. Front Immunol. 2024. PMID: 38873601 Free PMC article.
-
Whole-transcriptome analysis reveals the profiles and roles of coding and non-coding RNAs during hair follicle cycling in Rex rabbits.BMC Genomics. 2025 Jan 25;26(1):74. doi: 10.1186/s12864-025-11264-y. BMC Genomics. 2025. PMID: 39863835 Free PMC article.
-
Integrated analysis of lncRNAs and mRNAs by RNA-Seq in secondary hair follicle development and cycling (anagen, catagen and telogen) of Jiangnan cashmere goat (Capra hircus).BMC Vet Res. 2022 May 6;18(1):167. doi: 10.1186/s12917-022-03253-0. BMC Vet Res. 2022. PMID: 35524260 Free PMC article.
-
Integrative analysis reveals ncRNA-mediated molecular regulatory network driving secondary hair follicle regression in cashmere goats.BMC Genomics. 2018 Mar 27;19(1):222. doi: 10.1186/s12864-018-4603-3. BMC Genomics. 2018. PMID: 29587631 Free PMC article.
-
Construction of Potential Glioblastoma Multiforme-Related miRNA-mRNA Regulatory Network.Front Mol Neurosci. 2019 Mar 26;12:66. doi: 10.3389/fnmol.2019.00066. eCollection 2019. Front Mol Neurosci. 2019. PMID: 30971889 Free PMC article.
Cited by
-
Multi-omics and AI-driven advances in miRNA-mediated hair follicle regulation in cashmere goats.Front Vet Sci. 2025 Jul 9;12:1635202. doi: 10.3389/fvets.2025.1635202. eCollection 2025. Front Vet Sci. 2025. PMID: 40703926 Free PMC article. Review.
References
-
- Yang Q., Meng X., Xia L., Feng Z. Conservation status and causes of decline of musk deer (Moschus spp.) in China. Biol. Conserv. 2003;109:333–342. doi: 10.1016/S0006-3207(02)00159-3. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

