Sll1252 Coordinates Electron Transport between Plastoquinone and Cytochrome b6/f Complex in Synechocystis PCC 6803
- PMID: 38136973
- PMCID: PMC10743179
- DOI: 10.3390/genes14122151
Sll1252 Coordinates Electron Transport between Plastoquinone and Cytochrome b6/f Complex in Synechocystis PCC 6803
Abstract
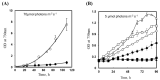
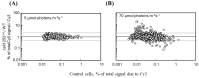
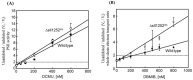
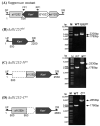
A mutant, Δsll1252ins, was generated to functionally characterize Sll1252. Δsll1252ins exhibited a slow-growth phenotype at 70 µmol photons m-2 s-1 and glucose sensitivity. In Δsll1252ins, the rate of PSII activity was not affected, whereas the whole chain electron transport activity was reduced by 45%. The inactivation of sll1252 led to the upregulation of genes, which were earlier reported to be induced in DBMIB-treated wild-type, suggesting that Sll1252 may be involved in electron transfer from the reduced-PQ pool to Cyt b6/f. The inhibitory effect of DCMU on PSII activity was similar in both wild-type and Δsll1252ins. However, the concentration of DBMIB for 50% inhibition of whole chain electron transport activity was 140 nM for Δsll1252ins and 300 nM for wild-type, confirming the site of action of Sll1252. Moreover, the elevated level of the reduced-PQ pool in Δsll1252ins supports that Sll1252 functions between the PQ pool and Cyt b6/f. Interestingly, we noticed that Δsll1252ins reverted to wild-type phenotype by insertion of natural transposon, ISY523, at the disruption site. Δsll1252-Ntrn, expressing only the C-terminal region of Sll1252, exhibited a slow-growth phenotype and disorganized thylakoid structure compared to wild-type and Δsll1252-Ctrn (expressing only the N-terminal region). Collectively, our data suggest that Sll1252 regulates electron transfer between the PQ pool and the Cyt b6/f complex in the linear photosynthetic electron transport chain via coordinated function of both the N- and C-terminal regions of Sll1252.
Keywords: PQ pool; Sll1252; photosynthetic electron transport; suppressor mutant.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


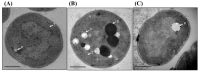
References
-
- Bhattacharya D., Medlin L. Phylogeny of plastids A review based on comparisons of small subunit ribosomal RNA coding regions. J. Phycol. 1995;31:487–496. doi: 10.1111/j.1529-8817.1995.tb02542.x. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

