Comparative Transcriptomic Assessment of Chemosensory Genes in Adult and Larval Olfactory Organs of Cnaphalocrocis medinalis
- PMID: 38136987
- PMCID: PMC10742765
- DOI: 10.3390/genes14122165
Comparative Transcriptomic Assessment of Chemosensory Genes in Adult and Larval Olfactory Organs of Cnaphalocrocis medinalis
Abstract
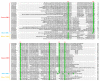
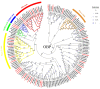
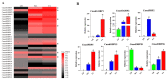
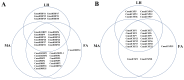
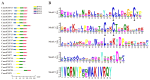
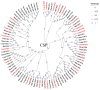
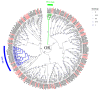
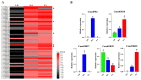
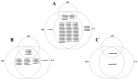
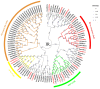
The rice leaf folder, Cnaphalocrocis medinalis (Lepidoptera: Pyralidae), is a notorious pest of rice in Asia. The larvae and adults of C. medinalis utilize specialized chemosensory systems to adapt to different environmental odors and physiological behaviors. However, the differences in chemosensory genes between the olfactory organs of these two different developmental stages remain unclear. Here, we conducted a transcriptome analysis of larvae heads, male antennae, and female antennae in C. medinalis and identified 131 putative chemosensory genes, including 32 OBPs (8 novel OBPs), 23 CSPs (2 novel CSPs), 55 ORs (17 novel ORs), 19 IRs (5 novel IRs) and 2 SNMPs. Comparisons between larvae and adults of C. medinalis by transcriptome and RT-qPCR analysis revealed that the number and expression of chemosensory genes in larval heads were less than that of adult antennae. Only 17 chemosensory genes (7 OBPs and 10 CSPs) were specifically or preferentially expressed in the larval heads, while a total of 101 chemosensory genes (21 OBPs, 9 CSPs, 51 ORs, 18 IRs, and 2 SNMPs) were specifically or preferentially expressed in adult antennae. Our study found differences in chemosensory gene expression between larvae and adults, suggesting their specialized functions at different developmental stages of C. medinalis. These results provide a theoretical basis for screening chemosensory genes as potential molecular targets and developing novel management strategies to control C. medinalis.
Keywords: Cnaphalocrocis medinalis; adult; chemosensory genes; expression pattern; larva; transcriptome analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Identification and Comparative Expression Profiles of Chemoreception Genes Revealed from Major Chemoreception Organs of the Rice Leaf Folder, Cnaphalocrocis medinalis (Lepidoptera: Pyralidae).PLoS One. 2015 Dec 11;10(12):e0144267. doi: 10.1371/journal.pone.0144267. eCollection 2015. PLoS One. 2015. PMID: 26657286 Free PMC article.
-
Analysis of a cDNA library from the antenna of Cnaphalocrocis medinalis and the expression pattern of olfactory genes.Biochem Biophys Res Commun. 2013 Apr 19;433(4):463-9. doi: 10.1016/j.bbrc.2013.03.038. Epub 2013 Mar 22. Biochem Biophys Res Commun. 2013. PMID: 23523786
-
Identification of Genes Putatively Involved in Chitin Metabolism and Insecticide Detoxification in the Rice Leaf Folder (Cnaphalocrocis medinalis) Larvae through Transcriptomic Analysis.Int J Mol Sci. 2015 Sep 10;16(9):21873-96. doi: 10.3390/ijms160921873. Int J Mol Sci. 2015. PMID: 26378520 Free PMC article.
-
Identification and Characterization of Candidate Chemosensory Gene Families from Spodoptera exigua Developmental Transcriptomes.Int J Biol Sci. 2015 Jul 15;11(9):1036-48. doi: 10.7150/ijbs.12020. eCollection 2015. Int J Biol Sci. 2015. PMID: 26221071 Free PMC article.
-
Identification of salivary proteins in the rice leaf folder Cnaphalocrocis medinalis by transcriptome and LC-MS/MS analyses.Insect Biochem Mol Biol. 2024 Nov;174:104191. doi: 10.1016/j.ibmb.2024.104191. Epub 2024 Oct 10. Insect Biochem Mol Biol. 2024. PMID: 39393440
Cited by
-
Editorial for the "Genetics, Phylogeny, and Evolution of Insects" Special Issue.Genes (Basel). 2024 Jul 30;15(8):1000. doi: 10.3390/genes15081000. Genes (Basel). 2024. PMID: 39202359 Free PMC article.
References
-
- Keil T.A. Insect Olfaction. Springer; Berlin, Germany: 1999. Morphology and development of the peripheral olfactory organs; pp. 5–47.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

