Unraveling Extremely Damaging IRAK4 Variants and Their Potential Implications for IRAK4 Inhibitor Efficacy
- PMID: 38138875
- PMCID: PMC10744719
- DOI: 10.3390/jpm13121648
Unraveling Extremely Damaging IRAK4 Variants and Their Potential Implications for IRAK4 Inhibitor Efficacy
Abstract
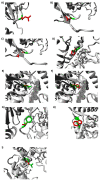
Interleukin-1-receptor-associated kinase 4 (IRAK4) possesses a crucial function in the toll-like receptor (TLR) signaling pathway, and the dysfunction of this molecule could lead to various infectious and immune-related diseases in addition to cancers. IRAK4 genetic variants have been linked to various types of diseases. Therefore, we conducted a comprehensive analysis to recognize the missense variants with the most damaging impacts on IRAK4 with the employment of diverse bioinformatics tools to study single-nucleotide polymorphisms' effects on function, stability, secondary structures, and 3D structure. The residues' location on the protein domain and their conservation status were investigated as well. Moreover, docking tools along with structural biology were engaged in analyzing the SNPs' effects on one of the developed IRAK4 inhibitors. By analyzing IRAK4 gene SNPs, the analysis distinguished ten variants as the most detrimental missense variants. All variants were situated in highly conserved positions on an important protein domain. L318S and L318F mutations were linked to changes in IRAK4 secondary structures. Eight SNPs were revealed to have a decreasing effect on the stability of IRAK4 via both I-Mutant 2.0 and Mu-Pro tools, while Mu-Pro tool identified a decreasing effect for the G198E SNP. In addition, detrimental effects on the 3D structure of IRAK4 were also discovered for the selected variants. Molecular modeling studies highlighted the detrimental impact of these identified SNP mutant residues on the druggability of the IRAK4 ATP-binding site towards the known target inhibitor, HG-12-6, as compared to the native protein. The loss of important ligand residue-wise contacts, altered protein global flexibility, increased steric clashes, and even electronic penalties at the ligand-binding site interfaces were all suggested to be associated with SNP models for hampering the HG-12-6 affinity towards IRAK4 target protein. This given model lays the foundation for the better prediction of various disorders relevant to IRAK4 malfunction and sheds light on the impact of deleterious IRAK4 variants on IRAK4 inhibitor efficacy.
Keywords: SNPs; TLR signaling; bioinformatics; innate immunity; molecular dynamics.
Conflict of interest statement
The authors declared no conflict of interest.
Figures





Similar articles
-
Design of Novel IRAK4 Inhibitors Using Molecular Docking, Dynamics Simulation and 3D-QSAR Studies.Molecules. 2022 Sep 24;27(19):6307. doi: 10.3390/molecules27196307. Molecules. 2022. PMID: 36234844 Free PMC article.
-
Conformational flexibility and inhibitor binding to unphosphorylated interleukin-1 receptor-associated kinase 4 (IRAK4).J Biol Chem. 2019 Mar 22;294(12):4511-4519. doi: 10.1074/jbc.RA118.005428. Epub 2019 Jan 24. J Biol Chem. 2019. PMID: 30679311 Free PMC article.
-
Mechanism of dysfunction of human variants of the IRAK4 kinase and a role for its kinase activity in interleukin-1 receptor signaling.J Biol Chem. 2018 Sep 28;293(39):15208-15220. doi: 10.1074/jbc.RA118.003831. Epub 2018 Aug 16. J Biol Chem. 2018. PMID: 30115681 Free PMC article.
-
Construction of IRAK4 inhibitor activity prediction model based on machine learning.Mol Divers. 2024 Aug;28(4):2289-2300. doi: 10.1007/s11030-024-10926-5. Epub 2024 Jul 6. Mol Divers. 2024. PMID: 38970641 Review.
-
Genetic and Epigenetic Regulation of the Innate Immune Response to Gout.Immunol Invest. 2023 Apr;52(3):364-397. doi: 10.1080/08820139.2023.2168554. Epub 2023 Feb 6. Immunol Invest. 2023. PMID: 36745138 Review.
Cited by
-
In vivo determination of analgesic and anti-inflammatory activities of isolated compounds from Cleome amblyocarpa and molecular modelling for the top active investigated compounds.RSC Adv. 2024 Aug 5;14(34):24503-24515. doi: 10.1039/d4ra04496g. eCollection 2024 Aug 5. RSC Adv. 2024. PMID: 39108954 Free PMC article.
References
-
- Sutherland A.M., Walley K.R., Nakada T.-A., Sham A.H.P., Wurfel M.M., Russell J.A. A Nonsynonymous Polymorphism of IRAK4 Associated with Increased Prevalence of Gram-Positive Infection and Decreased Response to Toll-like Receptor Ligands. J. Innate Immun. 2011;3:447–458. doi: 10.1159/000323880. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

