Genome-Wide Identification, Evolutionary Analysis, and Functional Studies of APX Genes in Melon (Cucuis melo L.)
- PMID: 38139399
- PMCID: PMC10743739
- DOI: 10.3390/ijms242417571
Genome-Wide Identification, Evolutionary Analysis, and Functional Studies of APX Genes in Melon (Cucuis melo L.)
Abstract
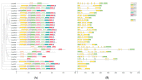
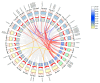
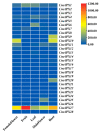
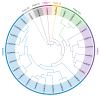
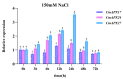
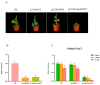
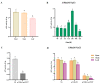
The antioxidative enzyme ascorbate peroxidase (APX) exerts a critically important function through scavenging reactive oxygen species (ROS), alleviating oxidative damage in plants, and enhancing their tolerance to salinity. Here, we identified 28 CmAPX genes that display an uneven distribution pattern throughout the 12 chromosomes of the melon genome by carrying out a bioinformatics analysis. Phylogenetic analyses revealed that the CmAPX gene family comprised seven different clades, with each clade of genes exhibiting comparable motifs and structures. We cloned 28 CmAPX genes to infer their encoded protein sequences; we then compared these sequences with proteins encoded by rice APX proteins (OsAPX2), Puccinellia tenuiflora APX proteins (PutAPX) and with pea APX proteins. We found that the CmAPX17, CmAPX24, and CmAPX27 genes in Clade I were closely related, and their structures were highly conserved. CmAPX27 (MELO3C020719.2.1) was found to promote resistance to 150 mM NaCl salt stress, according to quantitative real-time fluorescence PCR. Transcriptome data revealed that CmAPX27 was differentially expressed among tissues, and the observed differences in expression were significant. Virus-induced gene silencing of CmAPX27 significantly decreased salinity tolerance, and CmAPX27 exhibited differential expression in the leaf, stem, and root tissues of melon plants. This finding demonstrates that CmAPX27 exerts a key function in melon's tolerance to salt stress. Generally, CmAPX27 could be a target in molecular breeding efforts aimed at improving the salt tolerance of melon; further studies of CmAPX27 could unveil novel physiological mechanisms through which antioxidant enzymes mitigate the deleterious effects of ROS stress.
Keywords: APX gene family; functional validation; melon; salt stress.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


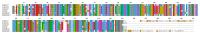
References
-
- Amanullah S., Gao P., Osae B.A., Saroj A., Yang T., Liu S., Weng Y., Luan F. Genetic linkage mapping and QTLs identification for morphology and fruit quality related traits of melon by SNP based CAPS markers. Sci. Hortic. 2021;278:109849. doi: 10.1016/j.scienta.2020.109849. - DOI
-
- Amanullah S., Liu S., Gao P., Zhu Z., Zhu Q., Fan C., Luan F. QTL mapping for melon (Cucumis melo L.) fruit traits by assembling and utilization of novel SNPs based CAPS markers. Sci. Hortic. 2018;236:18–29. doi: 10.1016/j.scienta.2018.02.041. - DOI
-
- Cabello M.J., Castellanos M.T., Romojaro F., Martínez-Madrid C., Ribas F. Yield and quality of melon grown under different irrigation and nitrogen rates. Agric. Water Manag. 2009;96:866–874. doi: 10.1016/j.agwat.2008.11.006. - DOI
-
- Rodríguez-López J.N., Espín J.C., Del Amor F., Tudela J., Martínez V., Cerdá A., García-Cánovas F. Purification and kinetic characterization of an anionic peroxidase from melon (Cucumis melo L.) cultivated under different salinity conditions. J. Agric. Food Chem. 2000;48:1537–1541. doi: 10.1021/jf9905774. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

