Characterization of Rotavirus Strains Responsible for Breakthrough Diarrheal Diseases among Zambian Children Using Whole Genome Sequencing
- PMID: 38140164
- PMCID: PMC10748035
- DOI: 10.3390/vaccines11121759
Characterization of Rotavirus Strains Responsible for Breakthrough Diarrheal Diseases among Zambian Children Using Whole Genome Sequencing
Abstract
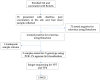
The occurrence of rotavirus (RV) infection among vaccinated children in high-burden settings poses a threat to further disease burden reduction. Genetically altered viruses have the potential to evade both natural infection and vaccine-induced immune responses, leading to diarrheal diseases among vaccinated children. Studies characterizing RV strains responsible for breakthrough infections in resource-limited countries where RV-associated diarrheal diseases are endemic are limited. We aimed to characterize RV strains detected in fully vaccinated children residing in Zambia using next-generation sequencing. We conducted whole genome sequencing on Illumina MiSeq. Whole genome assembly was performed using Geneious Prime 2023.1.2. A total of 76 diarrheal stool specimens were screened for RV, and 4/76 (5.2%) were RV-positive. Whole genome analysis revealed RVA/Human-wt/ZMB/CIDRZ-RV2088/2020/G1P[4]-I2-R2-C2-M2-A2-N2-T2-E2-H2 and RVA/Human-wt/ZMB/CIDRZ-RV2106/2020/G12P[4]-I1-R2-C2-M2-A2-N1-T2-E1-H2 strains were mono and multiple reassortant (exchanged genes in bold) respectively, whilst RVA/Human-wt/ZMB/CIDRZ-RV2150/2020/G12P[8]-I1-R1-C1-M1-A1-N1-T1-E1-H1 was a typical Wa-like strain. Comparison of VP7 and VP4 antigenic epitope of breakthrough strains and Rotarix strain revealed several amino acid differences. Variations in amino acids in antigenic epitope suggested they played a role in immune evasion of neutralizing antibodies elicited by vaccination. Findings from this study have the potential to inform national RV vaccination strategies and the design of highly efficacious universal RV vaccines.
Keywords: Zambia; breakthrough; children; reassortment; rotavirus; vaccine.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures




References
-
- Bagchi P., Dutta D., Chattopadhyay S., Mukherjee A., Halder U.C., Sarkar S., Kobayashi N., Komoto S., Taniguchi K., Chawla-Sarkar M. Rotavirus Nonstructural Protein 1 Suppresses Virus-Induced Cellular Apoptosis To Facilitate Viral Growth by Activating the Cell Survival Pathways during Early Stages of Infection. J. Virol. 2010;84:6834–6845. doi: 10.1128/JVI.00225-10. - DOI - PMC - PubMed
-
- Dóró R., László B., Martella V., Leshem E., Gentsch J., Parashar U., Bányai K. Review of global rotavirus strain prevalence data from six years post vaccine licensure surveillance: Is there evidence of strain selection from vaccine pressure? Infect. Genet. Evol. 2014;28:446–461. doi: 10.1016/j.meegid.2014.08.017. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

