COVID-19 Genomic Surveillance in Bangui (Central African Republic) Reveals a Landscape of Circulating Variants Linked to Validated Antiviral Targets of SARS-CoV-2 Proteome
- PMID: 38140550
- PMCID: PMC10748234
- DOI: 10.3390/v15122309
COVID-19 Genomic Surveillance in Bangui (Central African Republic) Reveals a Landscape of Circulating Variants Linked to Validated Antiviral Targets of SARS-CoV-2 Proteome
Abstract
Since its outbreak, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) spread rapidly, causing the Coronavirus Disease 19 (COVID-19) pandemic. Even with the vaccines' administration, the virus continued to circulate due to inequal access to prevention and therapeutic measures in African countries. Information about COVID-19 in Africa has been limited and contradictory, and thus regional studies are important. On this premise, we conducted a genomic surveillance study about COVID-19 lineages circulating in Bangui, Central African Republic (CAR). We collected 2687 nasopharyngeal samples at four checkpoints in Bangui from 2 to 22 July 2021. Fifty-three samples tested positive for SARS-CoV-2, and viral genomes were sequenced to look for the presence of different viral strains. We performed phylogenetic analysis and described the lineage landscape of SARS-CoV-2 circulating in the CAR along 15 months of pandemics and in Africa during the study period, finding the Delta variant as the predominant Variant of Concern (VoC). The deduced aminoacidic sequences of structural and non-structural genes were determined and compared to reference and reported isolates from Africa. Despite the limited number of positive samples obtained, this study provides valuable information about COVID-19 evolution at the regional level and allows for a better understanding of SARS-CoV-2 circulation in the CAR.
Keywords: COVID-19; Central African Republic; Delta variant; SARS-CoV-2; screening; severe acute respiratory syndrome coronavirus 2.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures




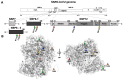

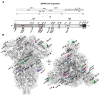
References
-
- COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU) [(accessed on 2 October 2023)]. Available online: https://coronavirus.jhu.edu/map.html.
-
- Robinson P.C., Liew D.F.L., Tanner H.L., Grainger J.R., Dwek R.A., Reisler R.B., Steinman L., Feldmann M., Ho L.P., Hussell T., et al. COVID-19 therapeutics: Challenges and directions for the future. Proc. Natl. Acad. Sci. USA. 2022;119:e2119893119. doi: 10.1073/pnas.2119893119. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

