Identification of Adenovirus E1B-55K Interaction Partners through a Common Binding Motif
- PMID: 38140597
- PMCID: PMC10747525
- DOI: 10.3390/v15122356
Identification of Adenovirus E1B-55K Interaction Partners through a Common Binding Motif
Abstract
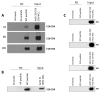
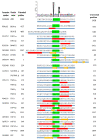
The adenovirus C5 E1B-55K protein is crucial for viral replication and is expressed early during infection. It can interact with E4orf6 to form a complex that functions as a ubiquitin E3 ligase. This complex targets specific cellular proteins and marks them for ubiquitination and, predominantly, subsequent proteasomal degradation. E1B-55K interacts with various proteins, with p53 being the most extensively studied, although identifying binding sites has been challenging. To explain the diverse range of proteins associated with E1B-55K, we hypothesized that other binding partners might recognize the simple p53 binding motif (xWxxxPx). In silico analyses showed that many known E1B-55K binding proteins possess this amino acid sequence; therefore, we investigated whether other xWxxxPx-containing proteins also bind to E1B-55K. Our findings revealed that many cellular proteins, including ATR, CHK1, USP9, and USP34, co-immunoprecipitate with E1B-55K. During adenovirus infection, several well-characterized E1B-55K binding proteins and newly identified interactors, including CSB, CHK1, and USP9, are degraded in a cullin-dependent manner. Notably, certain binding proteins, such as ATR and USP34, remain undegraded during infection. Structural predictions indicate no conservation of structure around the proposed binding motif, suggesting that the interaction relies on the correct arrangement of tryptophan and proline residues.
Keywords: MRE11; PR619; USP; adenovirus; cullin; early region 1B; p53; p53 binding motif.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

