Investigating Local Patterns of Mumps Virus Circulation, Using a Combination of Molecular Tools
- PMID: 38140661
- PMCID: PMC10747990
- DOI: 10.3390/v15122420
Investigating Local Patterns of Mumps Virus Circulation, Using a Combination of Molecular Tools
Abstract
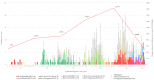
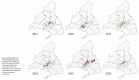
Mumps is a vaccine-preventable disease caused by the mumps virus (MuV). However, MuV has re-emerged in many countries with high vaccine coverage. The World Health Organization (WHO) recommends molecular surveillance based on sequencing of the small hydrophobic (SH) gene. Additionally, the combined use of SH and non-coding regions (NCR) has been described in different studies, proving to be a useful complement marker to discriminate general patterns of circulation at national and international levels. The aim of this work is to test local-level usefulness of the combination of SH and MF-NCR sequencing in tracing hidden transmission clusters and chains during the last epidemic wave (2015-2020) in Spain. A database with 903 cases from the Autonomous Community of Madrid was generated by the integration of microbiological and epidemiological data. Of these, 453 representative cases were genotyped. Eight different SH variants and thirty-four SH haplotypes were detected. Local MuV circulation showed the same temporal pattern previously described at a national level. Only two of the thirteen previously identified outbreaks were caused by more than one variant/haplotype. Geographical representation of SH variants allowed the identification of several previously undetected clusters, which were analysed phylogenetically by the combination of SH and MF-NCR, in a total of 90 cases. MF-NCR was not able to improve the discrimination of geographical clusters based on SH sequencing, showing limited resolution for outbreak investigations.
Keywords: MF-NCR sequence; SH sequence; genotyping; laboratory surveillance; molecular epidemiology; mumps; mumps virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Comparison of circulation patterns of mumps virus in the Netherlands and Spain (2015-2020).Front Microbiol. 2023 Jun 16;14:1207500. doi: 10.3389/fmicb.2023.1207500. eCollection 2023. Front Microbiol. 2023. PMID: 37396375 Free PMC article.
-
Genomic non-coding regions reveal hidden patterns of mumps virus circulation in Spain, 2005 to 2015.Euro Surveill. 2018 Apr;23(15):17-00349. doi: 10.2807/1560-7917.ES.2018.23.15.17-00349. Euro Surveill. 2018. PMID: 29667574 Free PMC article.
-
Increase of Diversity of Mumps Virus Genotype G SH Variants Circulating Among a Highly Immunized Population: Spain, 2007-2019.J Infect Dis. 2022 Dec 28;227(1):151-160. doi: 10.1093/infdis/jiac176. J Infect Dis. 2022. PMID: 35524966
-
The molecular epidemiology of mumps virus.Infect Genet Evol. 2004 Sep;4(3):215-9. doi: 10.1016/j.meegid.2004.02.003. Infect Genet Evol. 2004. PMID: 15450201 Review.
-
Genomic diversity of mumps virus and global distribution of the 12 genotypes.Rev Med Virol. 2015 Mar;25(2):85-101. doi: 10.1002/rmv.1819. Epub 2014 Nov 26. Rev Med Virol. 2015. PMID: 25424978 Review.
References
-
- Calendario Común de Vacunación a lo Largo de Toda la Vida. Consejo Interterritorial del Sistema Nacional de Salud, Ministerio de Sanidad; Madrid, Spain: 2023. pp. 2019–2021.
-
- De Frutos A.H., Olivares-Quintanar D., Masa-Calles J. Vigilancia epidemiológica y virológica de la parotiditis en España, 2005–2022. Epidemiological and virological surveillance of mumps. Boletín Epidemiológico. 2023;31:139–165. doi: 10.4321/s2173-92772023000300001. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

