Involvement of a Rarely Used Splicing SD2b Site in the Regulation of HIV-1 vif mRNA Production as Revealed by a Growth-Adaptive Mutation
- PMID: 38140666
- PMCID: PMC10747208
- DOI: 10.3390/v15122424
Involvement of a Rarely Used Splicing SD2b Site in the Regulation of HIV-1 vif mRNA Production as Revealed by a Growth-Adaptive Mutation
Abstract
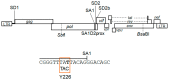
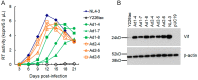
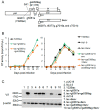
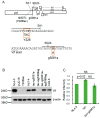
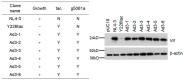
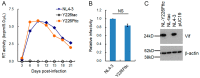
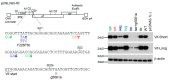
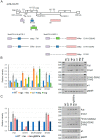
We have previously reported an HIV-1 mutant designated NL-Y226tac that expresses Vif at an ultra-low level, being replication-defective in high-APOBEC3G cells, such as H9. It carries a synonymous mutation within the splicing SA1 site relative to its parental clone. In order to determine whether a certain mutant(s) emerges during multi-infection cycles, we maintained H9 cells infected with a relatively low or high input of NL-Y226tac for extended time periods. Unexpectedly, we reproducibly identified a g5061a mutation in the SD2b site in the two independent long-term culture experiments that partially increases Vif expression and replication ability. Importantly, the adaptive mutation g5061a was demonstrated to enhance vif mRNA production by activation of the SA1 site mediated through increasing usage of a rarely used SD2b site. In the long-term culture initiated by a high virus input, we additionally found a Y226Fttc mutation at the original Y226tac site in SA1 that fully restores Vif expression and replication ability. As expected, the adaptive mutation Y226Fttc enhances vif mRNA production through increasing the splicing site usage of SA1. Our results here revealed the importance of the SD2b nucleotide sequence in producing vif mRNA involved in the HIV-1 adaptation and of mutual antagonism between Vif and APOBEC3 proteins in HIV-1 adaptation/evolution and survival.
Keywords: APOBEC3; HIV-1; SA1; SD2b; Vif; adaptation; splicing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
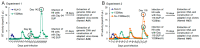
Similar articles
-
Regulation of Vif mRNA splicing by human immunodeficiency virus type 1 requires 5' splice site D2 and an exonic splicing enhancer to counteract cellular restriction factor APOBEC3G.J Virol. 2009 Jun;83(12):6067-78. doi: 10.1128/JVI.02231-08. Epub 2009 Apr 8. J Virol. 2009. PMID: 19357165 Free PMC article.
-
An intronic G run within HIV-1 intron 2 is critical for splicing regulation of vif mRNA.J Virol. 2013 Mar;87(5):2707-20. doi: 10.1128/JVI.02755-12. Epub 2012 Dec 19. J Virol. 2013. PMID: 23255806 Free PMC article.
-
A naturally occurring Vif mutant (I107T) attenuates anti-APOBEC3G activity and HIV-1 replication.J Mol Biol. 2013 Aug 23;425(16):2840-52. doi: 10.1016/j.jmb.2013.05.015. Epub 2013 May 23. J Mol Biol. 2013. PMID: 23707381
-
Various strategies for developing APOBEC3G protectors to circumvent human immunodeficiency virus type 1.Eur J Med Chem. 2023 Mar 15;250:115188. doi: 10.1016/j.ejmech.2023.115188. Epub 2023 Feb 6. Eur J Med Chem. 2023. PMID: 36773550 Review.
-
[Progress in the study of HIV-1 Vif and related inhibitors].Yao Xue Xue Bao. 2010 Jun;45(6):684-93. Yao Xue Xue Bao. 2010. PMID: 20939174 Review. Chinese.
Cited by
-
Interferon-Regulated Expression of Cellular Splicing Factors Modulates Multiple Levels of HIV-1 Gene Expression and Replication.Viruses. 2024 Jun 11;16(6):938. doi: 10.3390/v16060938. Viruses. 2024. PMID: 38932230 Free PMC article. Review.
References
-
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., COVID-19 Genomics UK (COG-UK) Consortium et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

