Structural mechanism of TRPV5 inhibition by econazole
- PMID: 38141613
- PMCID: PMC10872542
- DOI: 10.1016/j.str.2023.11.012
Structural mechanism of TRPV5 inhibition by econazole
Abstract
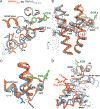
The calcium-selective TRPV5 channel activated by phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] is involved in calcium homeostasis. Recently, cryoelectron microscopy (cryo-EM) provided molecular details of TRPV5 modulation by exogenous and endogenous molecules. However, the details of TRPV5 inhibition by the antifungal agent econazole (ECN) remain elusive due to the low resolution of the currently available structure. In this study, we employ cryo-EM to comprehensively examine how the ECN inhibits TRPV5. By combining our structural findings with site-directed mutagenesis, calcium measurements, electrophysiology, and molecular dynamics simulations, we determined that residues F472 and L475 on the S4 helix, along with residue W495 on the S5 helix, collectively constitute the ECN-binding site. Additionally, the structure of TRPV5 in the presence of ECN and PI(4,5)P2, which does not show the bound activator, reveals a potential inhibition mechanism in which ECN competes with PI(4,5)P2, preventing the latter from binding, and ultimately pore closure.
Keywords: ECN; TRP; Transient receptor potential channel; activation by PI(4,5)P(2); cryo electron microscopy; cryo-EM; gating mechanism inhibition; inhibition by econazole.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare that they have no competing interests.
Figures


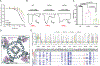
References
-
- Muller D, Hoenderop JG, Meij IC, van den Heuvel LP, Knoers NV, den Hollander AI, Eggert P, Garcia-Nieto V, Claverie-Martin F, and Bindels RJ (2000). Molecular cloning, tissue distribution, and chromosomal mapping of the human epithelial Ca2+ channel (ECAC1). Genomics 67, 48–53. 10.1006/geno.2000.6203. - DOI - PubMed
-
- van der Eerden BC, Hoenderop JG, de Vries TJ, Schoenmaker T, Buurman CJ, Uitterlinden AG, Pols HA, Bindels RJ, and van Leeuwen JP (2005). The epithelial Ca2+ channel TRPV5 is essential for proper osteoclastic bone resorption. Proc Natl Acad Sci U S A 102, 17507–17512. 10.1073/pnas.0505789102. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

