Targeting acid ceramidase enhances antitumor immune response in colorectal cancer
- PMID: 38142035
- PMCID: PMC11518951
- DOI: 10.1016/j.jare.2023.12.013
Targeting acid ceramidase enhances antitumor immune response in colorectal cancer
Abstract
Introduction: Acid ceramidase (hereafter referred as ASAH1) is an enzyme in sphingolipid metabolism that converts pro-survival ceramide into sphingosine. ASAH1 has been shown to be overexpressed in certain cancers. However, the role of ASAH1 in colorectal cancer still remain elusive.
Objective: The present study is aimed to understand how ASAH1 regulates colorectal cancer (CRC) progression and resistance to checkpoint inhibitor therapy.
Methods: Both pharmacological and genetic silencing of ASAH1 was used in the study. In vitro experiments were done on human and mouse CRC cell lines. The in vivo studies were conducted in NOD-SCID and BALB/c mice models. The combination of ASAH1 inhibitor and checkpoint inhibitor was tested using a syngeneic tumor model of CRC. Transcriptomic and metabolomic analyses were done to understand the effect of ASAH1 silencing.
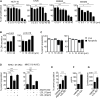
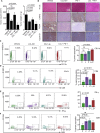
Results: ASAH1 is overexpressed in human CRC cases, and silencing the expression resulted in the induction of immunological cell death (ICD) and mitochondrial stress. The ASAH1 inhibitor (LCL-521), either as monotherapy or in combination with an anti-PD-1 antibody, resulted in reduction of tumors and, through induction of type I and II interferon response, activation of M1 macrophages and T cells, leading to enhanced infiltration of cytotoxic T cells. Our findings supported that the combination of LCL-521 and ICIs, which enhances the antitumor responses, and ASAH1 can be a druggable target in CRC.
Keywords: Acid ceramidase; Checkpoint inhibitor; Colorectal cancer; Immunological cell death; LCL-521; T cells.
Copyright © 2024. Production and hosting by Elsevier B.V.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures






References
-
- Aldea M., Andre F., Marabelle A., Dogan S., Barlesi F., Soria J.C. Overcoming resistance to tumor-targeted and immune-targeted therapies. Cancer Discov. 2021;11:874–899. - PubMed
-
- Coulie P.G., Van den Eynde B.J., van der Bruggen P., Boon T. Tumour antigens recognized by T lymphocytes: At the core of cancer immunotherapy. Nat Rev Cancer. 2014;14:135–146. - PubMed
-
- Brochier W., Bricard O., Coulie P.G. Facts and hopes in cancer antigens recognized by T cells. Clin Cancer Res. 2023;29:309–315. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

