Clearance of VWF by hepatic macrophages is critical for the protective effect of ADAMTS13 in sickle cell anemia mice
- PMID: 38142410
- PMCID: PMC10997916
- DOI: 10.1182/blood.2023021583
Clearance of VWF by hepatic macrophages is critical for the protective effect of ADAMTS13 in sickle cell anemia mice
Abstract
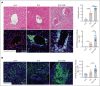
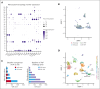
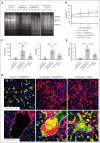
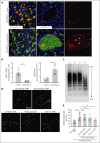
Although it is caused by a single-nucleotide mutation in the β-globin gene, sickle cell anemia (SCA) is a systemic disease with complex, incompletely elucidated pathologies. The mononuclear phagocyte system plays critical roles in SCA pathophysiology. However, how heterogeneous populations of hepatic macrophages contribute to SCA remains unclear. Using a combination of single-cell RNA sequencing and spatial transcriptomics via multiplexed error-robust fluorescence in situ hybridization, we identified distinct macrophage populations with diversified origins and biological functions in SCA mouse liver. We previously found that administering the von Willebrand factor (VWF)-cleaving protease ADAMTS13 alleviated vaso-occlusive episode in mice with SCA. Here, we discovered that the ADAMTS13-cleaved VWF was cleared from the circulation by a Clec4f+Marcohigh macrophage subset in a desialylation-dependent manner in the liver. In addition, sickle erythrocytes were phagocytized predominantly by Clec4f+Marcohigh macrophages. Depletion of macrophages not only abolished the protective effect of ADAMTS13 but exacerbated vaso-occlusive episode in mice with SCA. Furthermore, promoting macrophage-mediated VWF clearance reduced vaso-occlusion in SCA mice. Our study demonstrates that hepatic macrophages are important in the pathogenesis of SCA, and efficient clearance of VWF by hepatic macrophages is critical for the protective effect of ADAMTS13 in SCA mice.
© 2024 American Society of Hematology. Published by Elsevier Inc. All rights are reserved, including those for text and data mining, AI training, and similar technologies.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for B.S. is Lindsley F. Kimball Research Institute of New York Blood Center, New York, NY.
Figures






Comment in
-
Sickle cell anemia: hepatic macrophages to the rescue.Blood. 2024 Mar 28;143(13):1209-1210. doi: 10.1182/blood.2023023577. Blood. 2024. PMID: 38546640 No abstract available.
References
-
- Rees DC, Williams TN, Gladwin MT. Sickle-cell disease. Lancet. 2010;376(9757):2018–2031. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

