Whole-genome CRISPR screening identifies molecular mechanisms of PD-L1 expression in adult T-cell leukemia/lymphoma
- PMID: 38142436
- PMCID: PMC11033594
- DOI: 10.1182/blood.2023021423
Whole-genome CRISPR screening identifies molecular mechanisms of PD-L1 expression in adult T-cell leukemia/lymphoma
Abstract
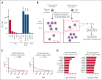
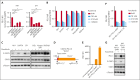
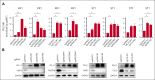
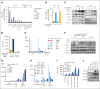
Adult T-cell leukemia/lymphoma (ATLL) is an aggressive T-cell malignancy with a poor prognosis and limited treatment options. Programmed cell death ligand 1(PD-L1) is recognized to be involved in the pathobiology of ATLL. However, what molecules control PD-L1 expression and whether genetic or pharmacological intervention might modify PD-L1 expression in ATLL cells are still unknown. To comprehend the regulatory mechanisms of PD-L1 expression in ATLL cells, we performed unbiased genome-wide clustered regularly interspaced short palindromic repeat (CRISPR) screening in this work. In ATLL cells, we discovered that the neddylation-associated genes NEDD8, NAE1, UBA3, and CUL3 negatively regulated PD-L1 expression, whereas STAT3 positively did so. We verified, in line with the genetic results, that treatment with the JAK1/2 inhibitor ruxolitinib or the neddylation pathway inhibitor pevonedistat resulted in a decrease in PD-L1 expression in ATLL cells or an increase in it, respectively. It is significant that these results held true regardless of whether ATLL cells had the PD-L1 3' structural variant, a known genetic anomaly that promotes PD-L1 overexpression in certain patients with primary ATLL. Pevonedistat alone showed cytotoxicity for ATLL cells, but compared with each single modality, pevonedistat improved the cytotoxic effects of the anti-PD-L1 monoclonal antibody avelumab and chimeric antigen receptor (CAR) T cells targeting PD-L1 in vitro. As a result, our work provided insight into a portion of the complex regulatory mechanisms governing PD-L1 expression in ATLL cells and demonstrated the in vitro preliminary preclinical efficacy of PD-L1-directed immunotherapies by using pevonedistat to upregulate PD-L1 in ATLL cells.
© 2024 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures


Comment in
-
Targeting PD-L1 to treat ATLL?Blood. 2024 Apr 4;143(14):1320-1322. doi: 10.1182/blood.2023023714. Blood. 2024. PMID: 38573604 No abstract available.
References
-
- Kataoka K, Shiraishi Y, Takeda Y, et al. Aberrant PD-L1 expression through 3'-UTR disruption in multiple cancers. Nature. 2016;534(7607):402–406. - PubMed
-
- Marcais A, Lhermitte L, Artesi M, et al. Targeted deep sequencing reveals clonal and subclonal mutational signatures in adult T-cell leukemia/lymphoma and defines an unfavorable indolent subtype. Leukemia. 2021;35(3):764–776. - PubMed
-
- Kataoka K, Nagata Y, Kitanaka A, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet. 2015;47(11):1304–1315. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

