The BRCA2 R2645G variant increases DNA binding and induces hyper-recombination
- PMID: 38142462
- PMCID: PMC11229362
- DOI: 10.1093/nar/gkad1222
The BRCA2 R2645G variant increases DNA binding and induces hyper-recombination
Erratum in
-
Correction to 'The BRCA2 R2645G variant increases DNA binding and induces hyper-recombination'.Nucleic Acids Res. 2024 Jul 8;52(12):7397. doi: 10.1093/nar/gkae482. Nucleic Acids Res. 2024. PMID: 38828779 Free PMC article. No abstract available.
Abstract
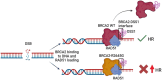
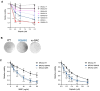
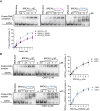
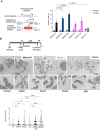
BRCA2 tumor suppressor protein ensures genome integrity by mediating DNA repair via homologous recombination (HR). This function is executed in part by its canonical DNA binding domain located at the C-terminus (BRCA2CTD), the only folded domain of the protein. Most germline pathogenic missense variants are located in this highly conserved region which binds to single-stranded DNA (ssDNA) and to the acidic protein DSS1. These interactions are essential for the HR function of BRCA2. Here, we report that the variant R2645G, identified in breast cancer and located at the DSS1 interface, unexpectedly increases the ssDNA binding activity of BRCA2CTDin vitro. Human cells expressing this variant display a hyper-recombination phenotype, chromosomal instability in the form of chromatid gaps when exposed to DNA damage, and increased PARP inhibitor sensitivity. In mouse embryonic stem cells (mES), this variant alters viability and confers sensitivity to cisplatin and Mitomycin C. These results suggest that BRCA2 interaction with ssDNA needs to be tightly regulated to limit HR and prevent chromosomal instability and we propose that this control mechanism involves DSS1. Given that several missense variants located within this region have been identified in breast cancer patients, these findings might have clinical implications for carriers.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Patel K.J., Yu V.P., Lee H., Corcoran A., Thistlethwaite F.C., Evans M.J., Colledge W.H., Friedman L.S., Ponder B.A., Venkitaraman A.R.. Involvement of Brca2 in DNA repair. Mol. Cell. 1998; 1:347–357. - PubMed
-
- Yang H., Jeffrey P.D., Miller J., Kinnucan E., Sun Y., Thoma N.H., Zheng N., Chen P.-L., Lee W.-H., Pavletich N.P.. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002; 297:1837–1848. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

