The TiHoCL panel for canine lymphoma: a feasibility study integrating functional genomics and network biology approaches for comparative oncology targeted NGS panel design
- PMID: 38144469
- PMCID: PMC10748409
- DOI: 10.3389/fvets.2023.1301536
The TiHoCL panel for canine lymphoma: a feasibility study integrating functional genomics and network biology approaches for comparative oncology targeted NGS panel design
Abstract
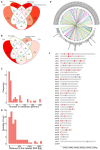
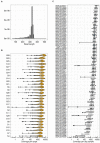
Targeted next-generation sequencing (NGS) enables the identification of genomic variants in cancer patients with high sensitivity at relatively low costs, and has thus opened the era to personalized human oncology. Veterinary medicine tends to adopt new technologies at a slower pace compared to human medicine due to lower funding, nonetheless it embraces technological advancements over time. Hence, it is reasonable to assume that targeted NGS will be incorporated into routine veterinary practice in the foreseeable future. Many animal diseases have well-researched human counterparts and hence, insights gained from the latter might, in principle, be harnessed to elucidate the former. Here, we present the TiHoCL targeted NGS panel as a proof of concept, exemplifying how functional genomics and network approaches can be effectively used to leverage the wealth of information available for human diseases in the development of targeted sequencing panels for veterinary medicine. Specifically, the TiHoCL targeted NGS panel is a molecular tool for characterizing and stratifying canine lymphoma (CL) patients designed based on human non-Hodgkin lymphoma (NHL) research outputs. While various single nucleotide polymorphisms (SNPs) have been associated with high risk of developing NHL, poor prognosis and resistance to treatment in NHL patients, little is known about the genetics of CL. Thus, the ~100 SNPs featured in the TiHoCL targeted NGS panel were selected using functional genomics and network approaches following a literature and database search that shielded ~500 SNPs associated with, in nearly all cases, human hematologic malignancies. The TiHoCL targeted NGS panel underwent technical validation and preliminary functional assessment by sequencing DNA samples isolated from blood of 29 lymphoma dogs using an Ion Torrent™ PGM System achieving good sequencing run metrics. Our design framework holds new possibilities for the design of similar molecular tools applied to other diseases for which limited knowledge is available and will improve drug target discovery and patient care.
Keywords: canine lymphoma; comparative genomics; comparative oncology; genomic variants; network biology; patient stratification; personalized oncology; targeted next-generation sequencing.
Copyright © 2023 Fibi-Smetana, Inglis, Schuster, Eberle, Granados-Soler, Liu, Krohn, Junghanss, Nolte, Taher and Murua Escobar.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Advancements in genetic analysis: Insights from a case study and review of next-generation sequencing techniques for veterinary oncology applications.Vet Clin Pathol. 2024 Oct 4. doi: 10.1111/vcp.13388. Online ahead of print. Vet Clin Pathol. 2024. PMID: 39367609 Review.
-
Canine Oncopanel: A capture-based, NGS platform for evaluating the mutational landscape and detecting putative driver mutations in canine cancers.Vet Comp Oncol. 2022 Mar;20(1):91-101. doi: 10.1111/vco.12746. Epub 2021 Aug 9. Vet Comp Oncol. 2022. PMID: 34286913
-
Development and analytical validation of a 25-gene next generation sequencing panel that includes the BRCA1 and BRCA2 genes to assess hereditary cancer risk.BMC Cancer. 2015 Apr 2;15:215. doi: 10.1186/s12885-015-1224-y. BMC Cancer. 2015. PMID: 25886519 Free PMC article.
-
LYmphoid NeXt-Generation Sequencing (LYNX) Panel: A Comprehensive Capture-Based Sequencing Tool for the Analysis of Prognostic and Predictive Markers in Lymphoid Malignancies.J Mol Diagn. 2021 Aug;23(8):959-974. doi: 10.1016/j.jmoldx.2021.05.007. Epub 2021 Jun 1. J Mol Diagn. 2021. PMID: 34082072
-
Applications and analysis of targeted genomic sequencing in cancer studies.Comput Struct Biotechnol J. 2019 Nov 7;17:1348-1359. doi: 10.1016/j.csbj.2019.10.004. eCollection 2019. Comput Struct Biotechnol J. 2019. PMID: 31762958 Free PMC article. Review.
Cited by
-
Application of Next-Generation Sequencing (NGS) Techniques for Selected Companion Animals.Animals (Basel). 2024 May 27;14(11):1578. doi: 10.3390/ani14111578. Animals (Basel). 2024. PMID: 38891625 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

