PanPA: generation and alignment of panproteome graphs
- PMID: 38145107
- PMCID: PMC10748787
- DOI: 10.1093/bioadv/vbad167
PanPA: generation and alignment of panproteome graphs
Abstract
Motivation: Compared to eukaryotes, prokaryote genomes are more diverse through different mechanisms, including a higher mutation rate and horizontal gene transfer. Therefore, using a linear representative reference can cause a reference bias. Graph-based pangenome methods have been developed to tackle this problem. However, comparisons in DNA space are still challenging due to this high diversity. In contrast, amino acid sequences have higher similarity due to evolutionary constraints, whereby a single amino acid may be encoded by several synonymous codons. Coding regions cover the majority of the genome in prokaryotes. Thus, panproteomes present an attractive alternative leveraging the higher sequence similarity while not losing much of the genome in non-coding regions.
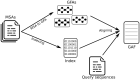
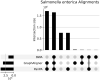
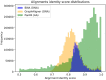
Results: We present PanPA, a method that takes a set of multiple sequence alignments of protein sequences, indexes them, and builds a graph for each multiple sequence alignment. In the querying step, it can align DNA or amino acid sequences back to these graphs. We first showcase that PanPA generates correct alignments on a panproteome from 1350 Escherichia coli. To demonstrate that panproteomes allow comparisons at longer phylogenetic distances, we compare DNA and protein alignments from 1073 Salmonella enterica assemblies against E.coli reference genome, pangenome, and panproteome using BWA, GraphAligner, and PanPA, respectively; with PanPA aligning around 22% more sequences. We also aligned a DNA short-reads whole genome sequencing (WGS) sample from S.enterica against the E.coli reference with BWA and the panproteome with PanPA, where PanPA was able to find alignment for 68% of the reads compared to 5% with BWA.
Availalability and implementation: PanPA is available at https://github.com/fawaz-dabbaghieh/PanPA.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures





References
-
- Akutsu T. A Linear Time Pattern Matching Algorithm Between a String and a Tree, Combinatorial Pattern Matching, Lecture Notes in Computer Science, Vol. 684, Springer-Verlag, Berlin/Heidelberg, 1993, 1–10.
-
- Amir A, Lewenstein M, Lewenstein N. et al. Pattern matching in hypertext. J Algorithms 2000;35:82–99.
LinkOut - more resources
Full Text Sources

