A workflow for the detection of antibiotic residues, measurement of water chemistry and preservation of hospital sink drain samples for metagenomic sequencing
- PMID: 38145816
- PMCID: PMC7617466
- DOI: 10.1016/j.jhin.2023.11.021
A workflow for the detection of antibiotic residues, measurement of water chemistry and preservation of hospital sink drain samples for metagenomic sequencing
Abstract
Background: Hospital sinks are environmental reservoirs that harbour healthcare-associated (HCA) pathogens. Selective pressures in sink environments, such as antibiotic residues, nutrient waste and hardness ions, may promote antibiotic resistance gene (ARG) exchange between bacteria. However, cheap and accurate sampling methods to characterize these factors are lacking.
Aims: To validate a workflow to detect antibiotic residues and evaluate water chemistry using dipsticks. Secondarily, to validate boric acid to preserve the taxonomic and ARG ('resistome') composition of sink trap samples for metagenomic sequencing.
Methods: Antibiotic residue dipsticks were validated against serial dilutions of ampicillin, doxycycline, sulfamethoxazole and ciprofloxacin, and water chemistry dipsticks against serial dilutions of chemical calibration standards. Sink trap aspirates were used for a 'real-world' pilot evaluation of dipsticks. To assess boric acid as a preservative of microbial diversity, the impact of incubation with and without boric acid at ∼22 °C on metagenomic sequencing outputs was evaluated at Day 2 and Day 5 compared with baseline (Day 0).
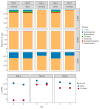
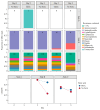
Findings: The limits of detection for each antibiotic were: 3 μg/L (ampicillin), 10 μg/L (doxycycline), 20 μg/L (sulfamethoxazole) and 8 μg/L (ciprofloxacin). The best performing water chemistry dipstick correctly characterized 34/40 (85%) standards in a concentration-dependent manner. One trap sample tested positive for the presence of tetracyclines and sulphonamides. Taxonomic and resistome composition were largely maintained after storage with boric acid at ∼22 °C for up to five days.
Conclusions: Dipsticks can be used to detect antibiotic residues and characterize water chemistry in sink trap samples. Boric acid was an effective preservative of trap sample composition, representing a low-cost alternative to cold-chain transport.
Keywords: Antibiotic residues; Antimicrobial resistance; Hospital sinks; Water chemistry.
Copyright © 2024 The Authors. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
All authors declare no conflicts of interest in this study.
Figures

Similar articles
-
The drainome: longitudinal metagenomic characterization of wastewater from hospital ward sinks to characterize the microbiome and resistome and to assess the effects of decontamination interventions.J Hosp Infect. 2024 Nov;153:55-62. doi: 10.1016/j.jhin.2024.06.005. Epub 2024 Jul 4. J Hosp Infect. 2024. PMID: 38969209 Free PMC article.
-
Survey of healthcare-associated sink infrastructure, and sink trap antibiotic residues and biochemistry, in twenty-nine UK hospitals.J Hosp Infect. 2025 May;159:140-147. doi: 10.1016/j.jhin.2025.02.002. Epub 2025 Feb 15. J Hosp Infect. 2025. PMID: 39961513
-
Water metagenomic analysis reveals low bacterial diversity and the presence of antimicrobial residues and resistance genes in a river containing wastewater from backyard aquacultures in the Mekong Delta, Vietnam.Environ Pollut. 2017 Mar;222:294-306. doi: 10.1016/j.envpol.2016.12.041. Epub 2017 Jan 3. Environ Pollut. 2017. PMID: 28062224
-
Exposure to doxycycline increases risk of carrying a broad range of enteric antimicrobial resistance determinants in an elderly cohort.J Infect. 2024 Oct;89(4):106243. doi: 10.1016/j.jinf.2024.106243. Epub 2024 Aug 12. J Infect. 2024. PMID: 39142392
-
Long-Term Exposure to Octenidine in a Simulated Sink Trap Environment Results in Selection of Pseudomonas aeruginosa, Citrobacter, and Enterobacter Isolates with Mutations in Efflux Pump Regulators.Appl Environ Microbiol. 2021 Apr 27;87(10):e00210-21. doi: 10.1128/AEM.00210-21. Print 2021 Apr 27. Appl Environ Microbiol. 2021. PMID: 33674437 Free PMC article.
Cited by
-
Divergent effects of azithromycin on purple corn (Zea mays L.) cultivation: Impact on biomass and antioxidant compounds.PLoS One. 2024 Aug 22;19(8):e0307548. doi: 10.1371/journal.pone.0307548. eCollection 2024. PLoS One. 2024. PMID: 39172948 Free PMC article.
-
The drainome: longitudinal metagenomic characterization of wastewater from hospital ward sinks to characterize the microbiome and resistome and to assess the effects of decontamination interventions.J Hosp Infect. 2024 Nov;153:55-62. doi: 10.1016/j.jhin.2024.06.005. Epub 2024 Jul 4. J Hosp Infect. 2024. PMID: 38969209 Free PMC article.
-
The hospital sink drain microbiome as a melting pot for AMR transmission to nosocomial pathogens.NPJ Antimicrob Resist. 2025 Jul 29;3(1):68. doi: 10.1038/s44259-025-00137-9. NPJ Antimicrob Resist. 2025. PMID: 40730864 Free PMC article. Review.
References
-
- World Health Organization. Antimicrobial Resistance. 2021. [last accessed June 2023]. Available at: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance.
-
- Hamers RL, Dobreva Z, Cassini A, Tamara A, Lazarus G, Asadinia KS, et al. Global knowledge gaps on antimicrobial resistance in the human health sector: a scoping review. Int J Infect Dis. 2023;134:142–9. - PubMed
-
- dos Santos LDR, Furlan JPR, Ramos MS, Gallo IFL, de Freitas LVP, Stehling EG. Co-occurrence of mcr-1, mcr-3, mcr-7 and clinically relevant antimicrobial resistance genes in environmental and fecal samples. Arch Microbiol. 2020;202:1795–800. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

