Formation of dsRNA by-products during in vitro transcription can be reduced by using low steady-state levels of UTP
- PMID: 38146535
- PMCID: PMC10749352
- DOI: 10.3389/fmolb.2023.1291045
Formation of dsRNA by-products during in vitro transcription can be reduced by using low steady-state levels of UTP
Abstract
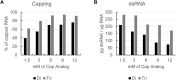
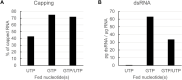
Introduction: Exogeneous messenger ribonucleic acid (mRNA) can be used as therapeutic and preventive medication. However, during the enzymatic production process, commonly called in vitro transcription, by-products occur which can reduce the therapeutic efficacy of mRNA. One such by-product is double-stranded RNA (dsRNA). We therefore sought to limit the generation of dsRNA by-products during in vitro transcription. Materials and methods: In vitro transcription was performed with a DNA template including a poly(A)-tail-encoding region, dinucleotide or trinucleotide cap analogs for cotranscriptional capping, and relevant nucleoside triphosphates. Concentrations of UTP or modified UTP (m1ΨTP) and GTP were reduced and fed over the course of the reaction. mRNA was analyzed for dsRNA contamination, yield of the reaction, RNA integrity, and capping efficiency before translational activity was assessed. Results: Limiting the steady-state level of UTP or m1ΨTP during the enzymatic reaction reduced dsRNA formation, while not affecting mRNA yield or RNA integrity. Capping efficiency was optimized with the use of a combined GTP and UTP or m1ΨTP feed, while still reducing dsRNA formation. Lower dsRNA levels led to higher protein expression from the corresponding mRNAs. Discussion: Low steady-state concentrations of UTP and GTP, fed in combination over the course of the in vitro transcription reaction, produce mRNA with high capping and low levels of dsRNA formation, resulting in high levels of protein expression. This novel approach may render laborious purification steps to remove dsRNA unnecessary.
Keywords: RNA capping; double-stranded RNA; in vitro transcription; mRNA translatability; mRNA-based therapeutics.
Copyright © 2023 Ziegenhals, Frieling, Wolf, Göbel, Koch, Lohmann, Baiersdörfer, Fesser, Sahin and Kuhn.
Conflict of interest statement
Authors TZ, RF, PW, KG, SK, ML, MB, SF, US, and AK were employed by BioNTech SE.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

