Aberrant monocytopoiesis drives granuloma development in sarcoidosis
- PMID: 38147536
- PMCID: PMC10935646
- DOI: 10.1093/intimm/dxad054
Aberrant monocytopoiesis drives granuloma development in sarcoidosis
Abstract
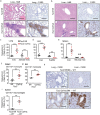
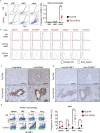
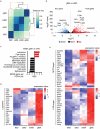
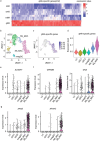
In sarcoidosis, granulomas develop in multiple organs including the liver and lungs. Although mechanistic target of rapamycin complex 1 (mTORC1) activation in macrophages drives granuloma development in sarcoidosis by enhancing macrophage proliferation, little is known about the macrophage subsets that proliferate and mature into granuloma macrophages. Here, we show that aberrantly increased monocytopoiesis gives rise to granulomas in a sarcoidosis model, in which Tsc2, a negative regulator of mTORC1, is conditionally deleted in CSF1R-expressing macrophages (Tsc2csf1rΔ mice). In Tsc2csf1rΔ mice, common myeloid progenitors (CMPs), granulocyte-monocyte progenitors (GMPs), common monocyte progenitors / monocyte progenitors (cMoPs / MPs), inducible monocyte progenitors (iMoPs), and Ly6Cint CX3CR1low CD14- immature monocytes (iMOs), but not monocyte-dendritic cell progenitors (MDPs) and common dendritic cell progenitors (CDPs), accumulated and proliferated in the spleen. Consistent with this, monocytes, neutrophils, and neutrophil-like monocytes increased in the spleens of Tsc2csf1rΔ mice, whereas dendritic cells did not. The adoptive transfer of splenic iMOs into wild-type mice gave rise to granulomas in the liver and lungs. In these target organs, iMOs matured into Ly6Chi classical monocytes/macrophages (cMOs). Giant macrophages (gMAs) also accumulated in the liver and lungs, which were similar to granuloma macrophages in expression of cell surface markers such as MerTK and SLAMF7. Furthermore, the gMA-specific genes were expressed in human macrophages from sarcoidosis skin lesions. These results suggest that mTORC1 drives granuloma development by promoting the proliferation of monocyte/neutrophil progenitors and iMOs predominantly in the spleen, and that proliferating iMOs mature into cMOs and then gMAs to give rise to granuloma after migration into the liver and lungs in sarcoidosis.
Keywords: mTORC1; macrophage.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Japanese Society for Immunology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures



Similar articles
-
Granulocyte-Monocyte Progenitors and Monocyte-Dendritic Cell Progenitors Independently Produce Functionally Distinct Monocytes.Immunity. 2017 Nov 21;47(5):890-902.e4. doi: 10.1016/j.immuni.2017.10.021. Immunity. 2017. PMID: 29166589 Free PMC article.
-
Aberrant Lipid Metabolism in Macrophages Is Associated with Granuloma Formation in Sarcoidosis.Am J Respir Crit Care Med. 2024 May 1;209(9):1152-1164. doi: 10.1164/rccm.202307-1273OC. Am J Respir Crit Care Med. 2024. PMID: 38353578 Free PMC article.
-
Chronic signaling via the metabolic checkpoint kinase mTORC1 induces macrophage granuloma formation and marks sarcoidosis progression.Nat Immunol. 2017 Mar;18(3):293-302. doi: 10.1038/ni.3655. Epub 2017 Jan 16. Nat Immunol. 2017. PMID: 28092373 Free PMC article.
-
Macrophage differentiation and granulomatous inflammation in osteopetrotic mice (op/op) defective in the production of CSF-1.Mol Reprod Dev. 1997 Jan;46(1):85-91. doi: 10.1002/(SICI)1098-2795(199701)46:1<85::AID-MRD13>3.0.CO;2-2. Mol Reprod Dev. 1997. PMID: 8981368 Review.
-
[Origin of epithelioid cells in sarcoid granuloma].Nihon Rinsho. 2002 Sep;60(9):1714-9. Nihon Rinsho. 2002. PMID: 12233065 Review. Japanese.
Cited by
-
mTORC1-Dependent Regulation of the CCL24-CCR3 Axis Controls Granuloma Formation and Maintenance in Sarcoidosis.bioRxiv [Preprint]. 2025 Jul 16:2025.07.11.664317. doi: 10.1101/2025.07.11.664317. bioRxiv. 2025. PMID: 40791394 Free PMC article. Preprint.
References
-
- Herrtwich L, Nanda I, Evangelou K, et al. . DNA damage signaling instructs polyploid macrophage fate in granulomas. Cell 2016;167:1264. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

