Tuning Potential Functions to Host-Guest Binding Data
- PMID: 38147689
- PMCID: PMC10838530
- DOI: 10.1021/acs.jctc.3c01050
Tuning Potential Functions to Host-Guest Binding Data
Abstract
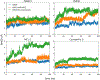
Software to more rapidly and accurately predict protein-ligand binding affinities is of high interest for early-stage drug discovery, and physics-based methods are among the most widely used technologies for this purpose. The accuracy of these methods depends critically on the accuracy of the potential functions that they use. Potential functions are typically trained against a combination of quantum chemical and experimental data. However, although binding affinities are among the most important quantities to predict, experimental binding affinities have not to date been integrated into the experimental data set used to train potential functions. In recent years, the use of host-guest complexes as simple and tractable models of binding thermodynamics has gained popularity due to their small size and simplicity, relative to protein-ligand systems. Host-guest complexes can also avoid ambiguities that arise in protein-ligand systems such as uncertain protonation states. Thus, experimental host-guest binding data are an appealing additional data type to integrate into the experimental data set used to optimize potential functions. Here, we report the extension of the Open Force Field Evaluator framework to enable the systematic calculation of host-guest binding free energies and their gradients with respect to force field parameters, coupled with the curation of 126 host-guest complexes with available experimental binding free energies. As an initial application of this novel infrastructure, we optimized generalized Born (GB) cavity radii for the OBC2 GB implicit solvent model against experimental data for 36 host-guest systems. This refitting led to a dramatic improvement in accuracy for both the training set and a separate test set with 90 additional host-guest systems. The optimized radii also showed encouraging transferability from host-guest systems to 59 protein-ligand systems. However, the new radii are significantly smaller than the baseline radii and lead to excessively favorable hydration free energies (HFEs). Thus, users of the OBC2 GB model currently may choose between GB cavity radii that yield more accurate binding affinities and GB cavity radii that yield more accurate HFEs. We suspect that achieving good accuracy on both will require more far-reaching adjustments to the GB model. We note that binding free-energy calculations using the OBC2 model in OpenMM gain about a 10× speedup relative to corresponding explicit solvent calculations, suggesting a future role for implicit solvent absolute binding free-energy (ABFE) calculations in virtual compound screening. This study proves the principle of using host-guest systems to train potential functions that are transferrable to protein-ligand systems and provides an infrastructure that enables a range of applications.
Figures
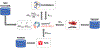

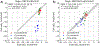
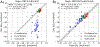


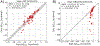
References
-
- Boresch S; Tettinger F; Leitgeb M; Karplus M Absolute Binding Free Energies: A Quantitative Approach for their Calculation. Journal of Physical Chemistry B 2003, 107, 9535–9551.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
