Morphomics via next-generation electron microscopy
- PMID: 38148118
- PMCID: PMC11167312
- DOI: 10.1093/jmcb/mjad081
Morphomics via next-generation electron microscopy
Abstract
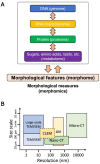
The living body is composed of innumerable fine and complex structures. Although these structures have been studied in the past, a vast amount of information pertaining to them still remains unknown. When attempting to observe these ultra-structures, the use of electron microscopy (EM) has become indispensable. However, conventional EM settings are limited to a narrow tissue area, which can bias observations. Recently, new trends in EM research have emerged, enabling coverage of far broader, nano-scale fields of view for two-dimensional wide areas and three-dimensional large volumes. Moreover, cutting-edge bioimage informatics conducted via deep learning has accelerated the quantification of complex morphological bioimages. Taken together, these technological and analytical advances have led to the comprehensive acquisition and quantification of cellular morphology, which now arises as a new omics science termed 'morphomics'.
Keywords: 3D bioimaging; comprehensive morphological analysis; deep learning; imaging database; next-generation electron microscopy.
© The Author(s) (2023). Published by Oxford University Press on behalf of Journal of Molecular Cell Biology, CEMCS, CAS.
Figures



References
-
- Barajas L. (1970). The ultrastructure of the juxtaglomerular apparatus as disclosed by three-dimensional reconstructions from serial sections. The anatomical relationship between the tubular and vascular components. J. Ultrastruct. Res. 33, 116–147. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

