Insights into Endophytic and Rhizospheric Bacteria of Five Sugar Beet Hybrids in Terms of Their Diversity, Plant-Growth Promoting, and Biocontrol Properties
- PMID: 38148389
- PMCID: PMC10751262
- DOI: 10.1007/s00248-023-02329-0
Insights into Endophytic and Rhizospheric Bacteria of Five Sugar Beet Hybrids in Terms of Their Diversity, Plant-Growth Promoting, and Biocontrol Properties
Abstract
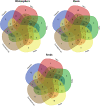
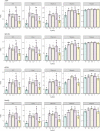
Sugar beet is the most important crop for sugar production in temperate zones. The plant microbiome is considered an important factor in crop productivity and health. Here, we investigated the bacterial diversity of seeds, roots, and rhizosphere of five sugar beet hybrids named Eduarda (ED), Koala (KO), Tibor (T), Tajfun (TF), and Cercospora-resistant (C). A culture-independent next-generation sequencing approach was used for the further investigation of seed-borne endophytes. Hybrid-associated bacteria were evaluated for their plant growth-promoting (PGP) characteristics, antagonistic activity towards Cercospora beticola and several Fusarium strains in dual culture assays, and drought and salinity tolerance. High-throughput sequencing revealed that the Proteobacteria phylum was most dominant in the seeds of all hybrids, followed by Cyanobacteria and Actinobacteriota. The predominant genus in all hybrids was Pantoea, followed by Pseudomonas, Acinetobacter, Chalicogloea, Corynebacterium, Enterobacter, Enterococcus, Glutamicibacter, Kosakonia, and Marinilactibacillus. Unique genera in the hybrids were Pleurocapsa and Arthrobacter (T), Klebsiella (TF), Apibacter (ED), and Alloscardovia (KO). The genera that were most represented in one hybrid were Weissella and Staphylococcus (TF); Streptococcus (T); Gardnerella, Prevotella, and Rothia (KO); and Gilliamella, Lactobacillus, and Snodgrassella (ED). Thirty-two bacteria out of 156 isolates from the rhizosphere, roots, and seeds were selected with respect to various plant growth-promoting activities in vitro, i.e., nitrogen fixation, phosphate solubilization, siderophore production, indole-3-acetic acid production, 1-aminocyclopropane-1-carboxylic acid deaminase activity, hydrogen cyanide production, exoenzymatic activity (amylase, protease, lipase, cellulase, xylanase, mannanases, gelatinase, and pectinase), mitigation of environmental stresses, and antifungal activity. Mixta theicola KO3-44, Providencia vermicola ED3-10, Curtobacterium pusillum ED2-6, and Bacillus subtilis KO3-18 had the highest potential to promote plant growth due to their multiple abilities (nitrogen fixation, phosphate solubilization, production of siderophores, and IAA). The best antagonistic activity towards phytopathogenic fungi was found for Bacillus velezensis C3-19, Paenibacillus polymyxa C3-36 and Bacillus halotolerans C3-16/2.1. Only four isolates B. velezensis T2-23, B. subtilis T3-4, B. velezensis ED2-2, and Bacillus halotolerans C3-16/2.1 all showed enzymatic activity, with the exception of xylanase production. B. halotolerans C3-16/2.1 exhibited the greatest tolerance to salinity, while two B. subtilis strains (C3-62 and TF2-1) grew successfully at the maximum concentration of PEG. The current study demonstrates that sugar beet-associated bacteria have a wide range of beneficial traits and are therefore highly promising for the formulation of biological control and PGP agents.
Keywords: Biotic and abiotic stresses; Cercospora-resistant hybrid; Keystone species; Plant-associated bacteria; Sugar beet hybrids.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

