Mitochondrial regulation of GPX4 inhibition-mediated ferroptosis in acute myeloid leukemia
- PMID: 38148395
- PMCID: PMC11082873
- DOI: 10.1038/s41375-023-02117-2
Mitochondrial regulation of GPX4 inhibition-mediated ferroptosis in acute myeloid leukemia
Erratum in
-
Correction: Mitochondrial regulation of GPX4 inhibition-mediated ferroptosis in acute myeloid leukemia.Leukemia. 2024 Apr;38(4):926. doi: 10.1038/s41375-024-02202-0. Leukemia. 2024. PMID: 38459170 No abstract available.
Abstract
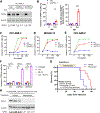
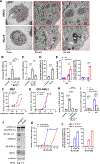
Resistance to apoptosis in acute myeloid leukemia (AML) cells causes refractory or relapsed disease, associated with dismal clinical outcomes. Ferroptosis, a mode of non-apoptotic cell death triggered by iron-dependent lipid peroxidation, has been investigated as potential therapeutic modality against therapy-resistant cancers, but our knowledge of its role in AML is limited. We investigated ferroptosis in AML cells and identified its mitochondrial regulation as a therapeutic vulnerability. GPX4 knockdown induced ferroptosis in AML cells, accompanied with characteristic mitochondrial lipid peroxidation, exerting anti-AML effects in vitro and in vivo. Electron transport chains (ETC) are primary sources of coenzyme Q10 (CoQ) recycling for its function of anti-lipid peroxidation in mitochondria. We found that the mitochondria-specific CoQ potently inhibited GPX4 inhibition-mediated ferroptosis, suggesting that mitochondrial lipid redox regulates ferroptosis in AML cells. Consistently, Rho0 cells, which lack functional ETC, were more sensitive to GPX4 inhibition-mediated mitochondrial lipid peroxidation and ferroptosis than control cells. Furthermore, degradation of ETC through hyperactivation of a mitochondrial protease, caseinolytic protease P (ClpP), synergistically enhanced the anti-AML effects of GPX4 inhibition. Collectively, our findings indicate that in AML cells, GPX4 inhibition induces ferroptosis, which is regulated by mitochondrial lipid redox and ETC.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
MA is a stockholder of Chimerix. JI, MA, and ADS have filed invention disclosure forms related to the use of imipridones in cancers. ADS has received research funding from Takeda Pharmaceuticals, BMS and Medivir AB, and consulting fees/honorarium from Takeda, Novartis, Jazz, and Otsuka Pharmaceuticals. ADS is named on a patent application for the use of DNT cells to treat AML. ADS is a member of the Medical and Scientific Advisory Board of the Leukemia and Lymphoma Society of Canada
The original online version of this article was revised: In this article the funding from M.L.P. 2nd was supported by the 1R25CA240137 UPWARDS Training Program and the CPRIT Research Training Award CPRIT Training Program (RP210028) was omitted.
Figures



References
-
- National Cancer Institute. Surveillance, Epidemiology, and End Results Program: Cancer Stat Facts: Leukemia — Acute Myeloid Leukemia (AML) 2022. [Available from: https://seer.cancer.gov/statfacts/html/amyl.html.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

