Epigenetic targeting of autophagy for cancer: DNA and RNA methylation
- PMID: 38148841
- PMCID: PMC10749975
- DOI: 10.3389/fonc.2023.1290330
Epigenetic targeting of autophagy for cancer: DNA and RNA methylation
Abstract
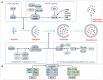
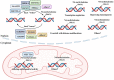
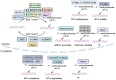
Autophagy, a crucial cellular mechanism responsible for degradation and recycling of intracellular components, is modulated by an intricate network of molecular signals. Its paradoxical involvement in oncogenesis, acting as both a tumor suppressor and promoter, has been underscored in recent studies. Central to this regulatory network are the epigenetic modifications of DNA and RNA methylation, notably the presence of N6-methyldeoxyadenosine (6mA) in genomic DNA and N6-methyladenosine (m6A) in eukaryotic mRNA. The 6mA modification in genomic DNA adds an extra dimension of epigenetic regulation, potentially impacting the transcriptional dynamics of genes linked to autophagy and, especially, cancer. Conversely, m6A modification, governed by methyltransferases and demethylases, influences mRNA stability, processing, and translation, affecting genes central to autophagic pathways. As we delve deeper into the complexities of autophagy regulation, the importance of these methylation modifications grows more evident. The interplay of 6mA, m6A, and autophagy points to a layered regulatory mechanism, illuminating cellular reactions to a range of conditions. This review delves into the nexus between DNA 6mA and RNA m6A methylation and their influence on autophagy in cancer contexts. By closely examining these epigenetic markers, we underscore their promise as therapeutic avenues, suggesting novel approaches for cancer intervention through autophagy modulation.
Keywords: 6mA methylation; autophagy; cancers; m6A methylation; therapy.
Copyright © 2023 Lin, Zhao, Zheng, Zhang, Li and Wu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Emerging Roles for DNA 6mA and RNA m6A Methylation in Mammalian Genome.Int J Mol Sci. 2023 Sep 9;24(18):13897. doi: 10.3390/ijms241813897. Int J Mol Sci. 2023. PMID: 37762200 Free PMC article. Review.
-
Epigenetic Methylations on N6-Adenine and N6-Adenosine with the same Input but Different Output.Int J Mol Sci. 2019 Jun 15;20(12):2931. doi: 10.3390/ijms20122931. Int J Mol Sci. 2019. PMID: 31208067 Free PMC article. Review.
-
Epigenetic N6-methyladenosine modification of RNA and DNA regulates cancer.Cancer Biol Med. 2020 Feb 15;17(1):9-19. doi: 10.20892/j.issn.2095-3941.2019.0347. Cancer Biol Med. 2020. PMID: 32296573 Free PMC article. Review.
-
Current insights into the implications of m6A RNA methylation and autophagy interaction in human diseases.Cell Biosci. 2021 Jul 27;11(1):147. doi: 10.1186/s13578-021-00661-x. Cell Biosci. 2021. PMID: 34315538 Free PMC article. Review.
-
Transcriptome-wide analysis of N6-methyladenosine uncovers its regulatory role in gene expression in the lepidopteran Bombyx mori.Insect Mol Biol. 2019 Oct;28(5):703-715. doi: 10.1111/imb.12584. Epub 2019 Apr 29. Insect Mol Biol. 2019. PMID: 30957943
Cited by
-
Nanomedicine-induced programmed cell death in cancer therapy: mechanisms and perspectives.Cell Death Discov. 2024 Aug 29;10(1):386. doi: 10.1038/s41420-024-02121-0. Cell Death Discov. 2024. PMID: 39209834 Free PMC article. Review.
-
Exploring the Role of mRNA Methylation in Insect Biology and Resistance.Insects. 2025 Apr 28;16(5):463. doi: 10.3390/insects16050463. Insects. 2025. PMID: 40429176 Free PMC article. Review.
-
Application of methylation in the diagnosis of ankylosing spondylitis.Clin Rheumatol. 2024 Oct;43(10):3073-3082. doi: 10.1007/s10067-024-07113-0. Epub 2024 Aug 21. Clin Rheumatol. 2024. PMID: 39167325 Review.
-
Molecular Mechanisms of Healthy Aging: The Role of Caloric Restriction, Intermittent Fasting, Mediterranean Diet, and Ketogenic Diet-A Scoping Review.Nutrients. 2024 Aug 28;16(17):2878. doi: 10.3390/nu16172878. Nutrients. 2024. PMID: 39275194 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

