ACE2 knockout hinders SARS-CoV-2 propagation in iPS cell-derived airway and alveolar epithelial cells
- PMID: 38149046
- PMCID: PMC10750251
- DOI: 10.3389/fcell.2023.1290876
ACE2 knockout hinders SARS-CoV-2 propagation in iPS cell-derived airway and alveolar epithelial cells
Erratum in
-
Corrigendum: ACE2 knockout hinders SARS-CoV-2 propagation in iPS cell-derived airway and alveolar epithelial cells.Front Cell Dev Biol. 2024 Jun 21;12:1407164. doi: 10.3389/fcell.2024.1407164. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38974145 Free PMC article.
Abstract
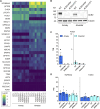
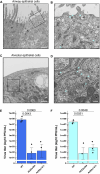
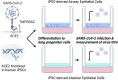
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), the causative agent of COVID-19, continues to spread around the world with serious cases and deaths. It has also been suggested that different genetic variants in the human genome affect both the susceptibility to infection and severity of disease in COVID-19 patients. Angiotensin-converting enzyme 2 (ACE2) has been identified as a cell surface receptor for SARS-CoV and SARS-CoV-2 entry into cells. The construction of an experimental model system using human iPS cells would enable further studies of the association between viral characteristics and genetic variants. Airway and alveolar epithelial cells are cell types of the lung that express high levels of ACE2 and are suitable for in vitro infection experiments. Here, we show that human iPS cell-derived airway and alveolar epithelial cells are highly susceptible to viral infection of SARS-CoV-2. Using gene knockout with CRISPR-Cas9 in human iPS cells we demonstrate that ACE2 plays an essential role in the airway and alveolar epithelial cell entry of SARS-CoV-2 in vitro. Replication of SARS-CoV-2 was strongly suppressed in ACE2 knockout (KO) lung cells. Our model system based on human iPS cell-derived lung cells may be applied to understand the molecular biology regulating viral respiratory infection leading to potential therapeutic developments for COVID-19 and the prevention of future pandemics.
Keywords: ACE2; CRISPR-Cas9; SARS-CoV-2; gene editing; gene knockout; iPS cells.
Copyright © 2023 Niwa, Sakai, Lung, Matsumoto, Mikawa, Maehana, Suzuki, Yamamoto, Maurissen, Hirabayashi, Noda, Kubo, Gotoh and Woltjen.
Conflict of interest statement
SG is a founder and stakeholder of HiLung, Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Corrigendum: ACE2 knockout hinders SARS-CoV-2 propagation in iPS cell-derived airway and alveolar epithelial cells.Front Cell Dev Biol. 2024 Jun 21;12:1407164. doi: 10.3389/fcell.2024.1407164. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38974145 Free PMC article.
-
Development of alveolar and airway cells from human iPS cells: toward SARS-CoV-2 research and drug toxicity testing.J Toxicol Sci. 2021;46(9):425-435. doi: 10.2131/jts.46.425. J Toxicol Sci. 2021. PMID: 34470994
-
Gene expression and in situ protein profiling of candidate SARS-CoV-2 receptors in human airway epithelial cells and lung tissue.Eur Respir J. 2020 Sep 3;56(3):2001123. doi: 10.1183/13993003.01123-2020. Print 2020 Sep. Eur Respir J. 2020. PMID: 32675206 Free PMC article.
-
SARS-CoV-2 pandemic and research gaps: Understanding SARS-CoV-2 interaction with the ACE2 receptor and implications for therapy.Theranostics. 2020 Jun 12;10(16):7448-7464. doi: 10.7150/thno.48076. eCollection 2020. Theranostics. 2020. PMID: 32642005 Free PMC article. Review.
-
Expression of ACE2 in airways: Implication for COVID-19 risk and disease management in patients with chronic inflammatory respiratory diseases.Clin Exp Allergy. 2020 Dec;50(12):1313-1324. doi: 10.1111/cea.13746. Epub 2020 Oct 6. Clin Exp Allergy. 2020. PMID: 32975865 Free PMC article. Review.
Cited by
-
Building a human lung from pluripotent stem cells to model respiratory viral infections.Respir Res. 2024 Jul 15;25(1):277. doi: 10.1186/s12931-024-02912-0. Respir Res. 2024. PMID: 39010108 Free PMC article. Review.
References
-
- Abo K. M., Sainz de Aja J., Lindstrom-Vautrin J., Alysandratos K. D., Richards A., Garcia-de-Alba C., et al. (2022). Air-liquid interface culture promotes maturation and allows environmental exposure of pluripotent stem cell–derived alveolar epithelium. JCI Insight 7 (6), e155589. 10.1172/jci.insight.155589 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

