Conditional in vivo deletion of LYN kinase has little effect on a BRCA1 loss-of-function-associated mammary tumour model
- PMID: 38149669
- PMCID: PMC10846530
- DOI: 10.1242/dmm.050211
Conditional in vivo deletion of LYN kinase has little effect on a BRCA1 loss-of-function-associated mammary tumour model
Abstract
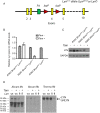
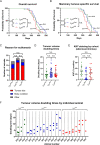
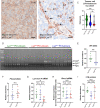
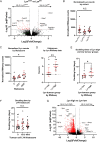
LYN kinase is expressed in BRCA1 loss-of-function-dependent mouse mammary tumours, in the cells of origin of such tumours, and in human breast cancer. Suppressing LYN kinase activity in BRCA1-defective cell lines as well as in in vitro cultures of Brca1-null mouse mammary tumours is deleterious to their growth. Here, we examined the interaction between LYN kinase and BRCA1 loss-of-function in an in vivo mouse mammary tumour model, using conditional knockout Brca1 and Lyn alleles. Comparison of Brca1 tumour cohorts showed little difference in mammary tumour formation between animals that were wild type, heterozygous or homozygous for the conditional Lyn allele, although this was confounded by factors including incomplete Lyn recombination in some tumours. RNA-sequencing analysis demonstrated that tumours with high levels of Lyn gene expression had a slower doubling time, but this was not correlated with levels of LYN staining in tumour cells themselves. Rather, high Lyn expression and slower tumour growth were likely a result of B-cell infiltration. The multifaceted role of LYN indicates that it is likely to present difficulties as a therapeutic target in breast cancer.
Keywords: BRCA1; Cre-LoxP; LYN kinase.
© 2024. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures

Similar articles
-
Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation.Nat Genet. 1999 May;22(1):37-43. doi: 10.1038/8743. Nat Genet. 1999. PMID: 10319859
-
Dissecting the heterogeneity and tumorigenesis of BRCA1 deficient mammary tumors via single cell RNA sequencing.Theranostics. 2021 Oct 25;11(20):9967-9987. doi: 10.7150/thno.63995. eCollection 2021. Theranostics. 2021. PMID: 34815798 Free PMC article.
-
The cell of origin of BRCA1 mutation-associated breast cancer: a cautionary tale of gene expression profiling.J Mammary Gland Biol Neoplasia. 2011 Apr;16(1):51-5. doi: 10.1007/s10911-011-9202-8. Epub 2011 Feb 19. J Mammary Gland Biol Neoplasia. 2011. PMID: 21336547 Review.
-
c-Kit is required for growth and survival of the cells of origin of Brca1-mutation-associated breast cancer.Oncogene. 2012 Feb 16;31(7):869-83. doi: 10.1038/onc.2011.289. Epub 2011 Jul 18. Oncogene. 2012. PMID: 21765473
-
Tumor formation in Brca1 conditional mutant mice.Environ Mol Mutagen. 2002;39(2-3):171-7. doi: 10.1002/em.10069. Environ Mol Mutagen. 2002. PMID: 11921186 Review.
References
-
- Buckley, N. E., Haddock, P., De Matos Simoes, R., Parkes, E., Irwin, G., Emmert-Streib, F., McQuaid, S., Kennedy, R. and Mullan, P. (2016). A BRCA1 deficient, NFκB driven immune signal predicts good outcome in triple negative breast cancer. Oncotarget 7, 19884-19896. 10.18632/oncotarget.7865 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

