Distinct Molecular Processes Mediate Donor-derived Cell-free DNA Release From Kidney Transplants in Different Disease States
- PMID: 38150492
- PMCID: PMC10962427
- DOI: 10.1097/TP.0000000000004877
Distinct Molecular Processes Mediate Donor-derived Cell-free DNA Release From Kidney Transplants in Different Disease States
Erratum in
-
Distinct Molecular Processes Mediate Donor-derived Cell-free DNA Release From Kidney Transplants in Different Disease States: Erratum.Transplantation. 2024 Apr 1;108(4):e68. doi: 10.1097/TP.0000000000004989. Epub 2024 Mar 23. Transplantation. 2024. PMID: 38526433 Free PMC article. No abstract available.
Abstract
Background: Among all biopsies in the Trifecta-Kidney Study ( ClinicalTrials.gov NCT04239703), elevated plasma donor-derived cell-free DNA (dd-cfDNA) correlated most strongly with molecular antibody-mediated rejection (AMR) but was also elevated in other states: T cell-mediated rejection (TCMR), acute kidney injury (AKI), and some apparently normal biopsies. The present study aimed to define the molecular correlates of plasma dd-cfDNA within specific states.
Methods: Dd-cfDNA was measured by the Prospera test. Molecular rejection and injury states were defined using the Molecular Microscope system. We studied the correlation between dd-cfDNA and the expression of genes, transcript sets, and classifier scores within specific disease states, and compared AMR, TCMR, and AKI to biopsies classified as normal and no injury (NRNI).
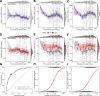
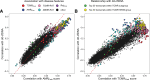
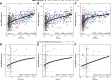
Results: In all 604 biopsies, dd-cfDNA was elevated in AMR, TCMR, and AKI. Within AMR biopsies, dd-cfDNA correlated with AMR activity and stage. Within AKI, the correlations reflected acute parenchymal injury, including cell cycling. Within biopsies classified as MMDx Normal and archetypal No injury (NRNI), dd-cfDNA still correlated significantly with rejection- and injury-related genes. TCMR activity (eg, the TCMR Prob classifier) correlated with dd-cfDNA, but within TCMR biopsies, top gene correlations were complex and not the top TCMR-selective genes.
Conclusions: In kidney transplants, elevated plasma dd-cfDNA is associated with 3 distinct molecular states in the donor tissue: AMR, recent parenchymal injury (including cell cycling), and TCMR, potentially complicated by parenchymal disruption. Moreover, subtle rejection- and injury-related changes in the donor tissue can contribute to dd-cfDNA elevations in transplants considered to have no rejection or injury.
Copyright © 2023 The Author(s). Published by Wolters Kluwer Health, Inc.
Conflict of interest statement
P.F.H. has shares in Transcriptome Sciences Inc (TSI), a University of Alberta research company with an interest in molecular diagnostics, and is a consultant to Natera Inc. All Natera Inc authors are employees and own equity at Natera Inc. The other authors declare no conflicts of interest.
Figures


References
-
- Moinuddin I, Kumar D, Kamal L, et al. Calibration of donor-derived cell-free DNA criteria for rejection with molecular diagnoses of kidney transplant biopsies. Am J Transplant. 2020;20:680–680.
-
- Huang E, Sethi S, Peng A, et al. Early clinical experience using donor-derived cell-free DNA to detect rejection in kidney transplant recipients. Am J Transplant. 2019;19:1663–1670. - PubMed
-
- Dengu F. Next-generation sequencing methods to detect donor-derived cell-free DNA after transplantation. Transplant Rev (Orlando). 2020;34:100542. - PubMed
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical

