Impact of various high fat diets on gene expression and the microbiome across the mouse intestines
- PMID: 38151490
- PMCID: PMC10752901
- DOI: 10.1038/s41598-023-49555-7
Impact of various high fat diets on gene expression and the microbiome across the mouse intestines
Abstract
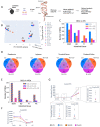
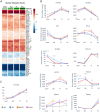
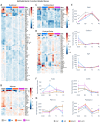
High fat diets (HFDs) have been linked to several diseases including obesity, diabetes, fatty liver, inflammatory bowel disease (IBD) and colon cancer. In this study, we examined the impact on intestinal gene expression of three isocaloric HFDs that differed only in their fatty acid composition-coconut oil (saturated fats), conventional soybean oil (polyunsaturated fats) and a genetically modified soybean oil (monounsaturated fats). Four functionally distinct segments of the mouse intestinal tract were analyzed using RNA-seq-duodenum, jejunum, terminal ileum and proximal colon. We found considerable dysregulation of genes in multiple tissues with the different diets, including those encoding nuclear receptors and genes involved in xenobiotic and drug metabolism, epithelial barrier function, IBD and colon cancer as well as genes associated with the microbiome and COVID-19. Network analysis shows that genes involved in metabolism tend to be upregulated by the HFDs while genes related to the immune system are downregulated; neurotransmitter signaling was also dysregulated by the HFDs. Genomic sequencing also revealed a microbiome altered by the HFDs. This study highlights the potential impact of different HFDs on gut health with implications for the organism as a whole and will serve as a reference for gene expression along the length of the intestines.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Update of
-
Impact of Various High Fat Diets on Gene Expression and the Microbiome Across the Mouse Intestines.Res Sq [Preprint]. 2023 Oct 9:rs.3.rs-3401763. doi: 10.21203/rs.3.rs-3401763/v1. Res Sq. 2023. Update in: Sci Rep. 2023 Dec 27;13(1):22758. doi: 10.1038/s41598-023-49555-7. PMID: 37886485 Free PMC article. Updated. Preprint.
Similar articles
-
Impact of Various High Fat Diets on Gene Expression and the Microbiome Across the Mouse Intestines.Res Sq [Preprint]. 2023 Oct 9:rs.3.rs-3401763. doi: 10.21203/rs.3.rs-3401763/v1. Res Sq. 2023. Update in: Sci Rep. 2023 Dec 27;13(1):22758. doi: 10.1038/s41598-023-49555-7. PMID: 37886485 Free PMC article. Updated. Preprint.
-
Gut Mucosal Proteins and Bacteriome Are Shaped by the Saturation Index of Dietary Lipids.Nutrients. 2019 Feb 16;11(2):418. doi: 10.3390/nu11020418. Nutrients. 2019. PMID: 30781503 Free PMC article.
-
Differential effects of coconut versus soy oil on gut microbiota composition and predicted metabolic function in adult mice.BMC Genomics. 2018 Nov 7;19(1):808. doi: 10.1186/s12864-018-5202-z. BMC Genomics. 2018. PMID: 30404613 Free PMC article.
-
Dietary Fat and Cancer-Which Is Good, Which Is Bad, and the Body of Evidence.Int J Mol Sci. 2020 Jun 9;21(11):4114. doi: 10.3390/ijms21114114. Int J Mol Sci. 2020. PMID: 32526973 Free PMC article. Review.
-
Negative Effects of a High-Fat Diet on Intestinal Permeability: A Review.Adv Nutr. 2020 Jan 1;11(1):77-91. doi: 10.1093/advances/nmz061. Adv Nutr. 2020. PMID: 31268137 Free PMC article. Review.
Cited by
-
Distinct gut flora profile induced by postnatal trans-fat diet in gestationally bisphenol A-exposed rats.PLoS One. 2024 Jul 9;19(7):e0306741. doi: 10.1371/journal.pone.0306741. eCollection 2024. PLoS One. 2024. PMID: 38980850 Free PMC article.
-
Programmable probiotic consortium employ an oleic acid-inducible system to sense and degrade cholesterol in high-fat diet mice.Gut Microbes. 2025 Dec;17(1):2531198. doi: 10.1080/19490976.2025.2531198. Epub 2025 Jul 12. Gut Microbes. 2025. PMID: 40650420 Free PMC article.
-
Brain-Gut and Microbiota-Gut-Brain Communication in Type-2 Diabetes Linked Alzheimer's Disease.Nutrients. 2024 Aug 3;16(15):2558. doi: 10.3390/nu16152558. Nutrients. 2024. PMID: 39125436 Free PMC article. Review.
-
Editorial: Hepatocyte nuclear factor 4 alpha - new insights into an old receptor.Front Endocrinol (Lausanne). 2024 Sep 25;15:1491965. doi: 10.3389/fendo.2024.1491965. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 39387053 Free PMC article. No abstract available.
-
Supplementation of a High-Fat Diet with Pentadecylresorcinol Increases the Representation of Akkermansia muciniphila in the Mouse Small and Large Intestines and May Protect against Complications Caused by Imbalanced Nutrition.Int J Mol Sci. 2024 Jun 15;25(12):6611. doi: 10.3390/ijms25126611. Int J Mol Sci. 2024. PMID: 38928317 Free PMC article.

