Protein N-myristoylation plays a critical role in the mitochondrial localization of human mitochondrial complex I accessory subunit NDUFB7
- PMID: 38151566
- PMCID: PMC10752898
- DOI: 10.1038/s41598-023-50390-z
Protein N-myristoylation plays a critical role in the mitochondrial localization of human mitochondrial complex I accessory subunit NDUFB7
Abstract
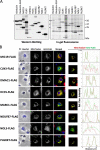
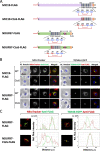
The present study examined human N-myristoylated proteins that specifically localize to mitochondria among the 1,705 human genes listed in MitoProteome, a mitochondrial protein database. We herein employed a strategy utilizing cellular metabolic labeling with a bioorthogonal myristic acid analog in transfected COS-1 cells established in our previous studies. Four proteins, DMAC1, HCCS, NDUFB7, and PLGRKT, were identified as N-myristoylated proteins that specifically localize to mitochondria. Among these proteins, DMAC1 and NDUFB7 play critical roles in the assembly of complex I of the mitochondrial respiratory chain. DMAC1 functions as an assembly factor, and NDUFB7 is an accessory subunit of complex I. An analysis of the intracellular localization of non-myristoylatable G2A mutants revealed that protein N-myristoylation occurring on NDUFB7 was important for the mitochondrial localization of this protein. Furthermore, an analysis of the role of the CHCH domain in NDUFB7 using Cys to Ser mutants revealed that it was essential for the mitochondrial localization of NDUFB7. Therefore, the present results showed that NDUFB7, a vital component of human mitochondrial complex I, was N-myristoylated, and protein N-myrisotylation and the CHCH domain were both indispensable for the specific targeting and localization of NDUFB7 to mitochondria.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Identification and characterization of protein N-myristoylation occurring on four human mitochondrial proteins, SAMM50, TOMM40, MIC19, and MIC25.PLoS One. 2018 Nov 14;13(11):e0206355. doi: 10.1371/journal.pone.0206355. eCollection 2018. PLoS One. 2018. PMID: 30427857 Free PMC article.
-
Targeting and import mechanism of coiled-coil helix coiled-coil helix domain-containing protein 3 (ChChd3) into the mitochondrial intermembrane space.J Biol Chem. 2012 Nov 16;287(47):39480-91. doi: 10.1074/jbc.M112.387696. Epub 2012 Sep 27. J Biol Chem. 2012. PMID: 23019327 Free PMC article.
-
Identification of Human N-Myristoylated Proteins from Human Complementary DNA Resources by Cell-Free and Cellular Metabolic Labeling Analyses.PLoS One. 2015 Aug 26;10(8):e0136360. doi: 10.1371/journal.pone.0136360. eCollection 2015. PLoS One. 2015. PMID: 26308446 Free PMC article.
-
Protein N-myristoylation: critical role in apoptosis and salt tolerance.Sci STKE. 2000 Dec 19;2000(63):pe1. doi: 10.1126/stke.2000.63.pe1. Sci STKE. 2000. PMID: 11752628 Review.
-
Post-translational myristoylation: Fat matters in cellular life and death.Biochimie. 2011 Jan;93(1):18-31. doi: 10.1016/j.biochi.2010.10.018. Epub 2010 Nov 5. Biochimie. 2011. PMID: 21056615 Review.
Cited by
-
Comparative study of the three-dimensional genomes of granulosa cells in germinal vesicle and metaphase II follicles.Front Genet. 2024 Nov 20;15:1480153. doi: 10.3389/fgene.2024.1480153. eCollection 2024. Front Genet. 2024. PMID: 39634272 Free PMC article.
-
Protein lipidation in the tumor microenvironment: enzymology, signaling pathways, and therapeutics.Mol Cancer. 2025 May 7;24(1):138. doi: 10.1186/s12943-025-02309-7. Mol Cancer. 2025. PMID: 40335986 Free PMC article. Review.
-
Oxidative protein folding in the intermembrane space of human mitochondria.FEBS Open Bio. 2024 Oct;14(10):1610-1626. doi: 10.1002/2211-5463.13839. Epub 2024 Jun 12. FEBS Open Bio. 2024. PMID: 38867508 Free PMC article. Review.
-
Regulation of BCR-dependent germinal center B-cell formation by HGAL and insight into its emerging myeloid ortholog, C1ORF150.Front Immunol. 2024 Oct 15;15:1437516. doi: 10.3389/fimmu.2024.1437516. eCollection 2024. Front Immunol. 2024. PMID: 39474423 Free PMC article. Review.
-
Exploration of degrons and their ability to mediate targeted protein degradation.RSC Med Chem. 2025 Jan 1. doi: 10.1039/d4md00787e. Online ahead of print. RSC Med Chem. 2025. PMID: 39867589 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

