LNK/ SH2B3 as a novel driver in juvenile myelomonocytic leukemia
- PMID: 38152053
- PMCID: PMC11290546
- DOI: 10.3324/haematol.2023.283776
LNK/ SH2B3 as a novel driver in juvenile myelomonocytic leukemia
Abstract
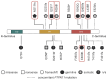
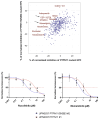
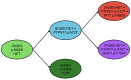
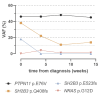
Mutations in five canonical Ras pathway genes (NF1, NRAS, KRAS, PTPN11 and CBL) are detected in nearly 90% of patients with juvenile myelomonocytic leukemia (JMML), a frequently fatal malignant neoplasm of early childhood. In this report, we describe seven patients diagnosed with SH2B3-mutated JMML, including five patients who were found to have initiating, loss-of-function mutations in the gene. SH2B3 encodes the adaptor protein LNK, a negative regulator of normal hematopoiesis upstream of the Ras pathway. These mutations were identified to be germline, somatic or a combination of both. Loss of function of LNK, which has been observed in other myeloid malignancies, results in abnormal proliferation of hematopoietic cells due to cytokine hypersensitivity and activation of the JAK/STAT signaling pathway. In vitro studies of induced pluripotent stem cell-derived JMML-like hematopoietic progenitor cells also demonstrated sensitivity of SH2B3-mutated hematopoietic progenitor cells to JAK inhibition. Lastly, we describe two patients with JMML and SH2B3 mutations who were treated with the JAK1/2 inhibitor ruxolitinib. This report expands the spectrum of initiating mutations in JMML and raises the possibility of targeting the JAK/STAT pathway in patients with SH2B3 mutations.
Figures
Comment in
-
SH2B3 alterations in a novel genetic condition, juvenile myelomonocytic leukemia, and myeloproliferative neoplasia.Haematologica. 2024 Aug 1;109(8):2391-2394. doi: 10.3324/haematol.2023.284747. Haematologica. 2024. PMID: 38618667 Free PMC article. No abstract available.
Similar articles
-
Germline bi-allelic SH2B3/LNK alteration predisposes to a neonatal juvenile myelomonocytic leukemia-like disorder.Haematologica. 2024 Aug 1;109(8):2542-2554. doi: 10.3324/haematol.2023.283917. Haematologica. 2024. PMID: 37981895 Free PMC article.
-
RAS Pathway Mutation Patterns in Patients With Juvenile Myelomonocytic Leukemia: A Developing Country Single-center Experience.Clin Lymphoma Myeloma Leuk. 2020 Jul;20(7):e368-e374. doi: 10.1016/j.clml.2020.02.003. Epub 2020 Feb 12. Clin Lymphoma Myeloma Leuk. 2020. PMID: 32209330
-
Integrated molecular profiling of juvenile myelomonocytic leukemia.Blood. 2018 Apr 5;131(14):1576-1586. doi: 10.1182/blood-2017-07-798157. Epub 2018 Feb 2. Blood. 2018. PMID: 29437595
-
JMML genomics and decisions.Hematology Am Soc Hematol Educ Program. 2018 Nov 30;2018(1):307-312. doi: 10.1182/asheducation-2018.1.307. Hematology Am Soc Hematol Educ Program. 2018. PMID: 30504325 Free PMC article. Review.
-
Targeted therapy in juvenile myelomonocytic leukemia: Where are we now?Pediatr Blood Cancer. 2022 Nov;69(11):e29930. doi: 10.1002/pbc.29930. Epub 2022 Sep 12. Pediatr Blood Cancer. 2022. PMID: 36094370 Review.
Cited by
-
Update on Recommendations for Surveillance for Children with Predisposition to Hematopoietic Malignancy.Clin Cancer Res. 2024 Oct 1;30(19):4286-4295. doi: 10.1158/1078-0432.CCR-24-0685. Clin Cancer Res. 2024. PMID: 39078402 Free PMC article. Review.
-
SH2B3 alterations in a novel genetic condition, juvenile myelomonocytic leukemia, and myeloproliferative neoplasia.Haematologica. 2024 Aug 1;109(8):2391-2394. doi: 10.3324/haematol.2023.284747. Haematologica. 2024. PMID: 38618667 Free PMC article. No abstract available.
References
-
- Locatelli F, Niemeyer CM. How I treat juvenile myelomonocytic leukemia. Blood. 2018;125(7):1083-1091. - PubMed
-
- Emanuel PD, Bates LJ, Castleberry RP, Gualtieri RJ, Zuckerman KS. Selective hypersensitivity to granulocyte-macrophage colony-stimulating factor by juvenile chronic myeloid leukemia hematopoietic progenitors. Blood. 1991;77(5):925-929. - PubMed
-
- Caye A, Strullu M, Guidez F, et al. . Juvenile myelomonocytic leukemia displays mutations in components of the RAS pathway and the PRC2 network. Nat Genet. 2015;47(11):1334-1340. - PubMed
-
- Murakami N, Okuno Y, Yoshida K, et al. . Integrated molecular profiling of juvenile myelomonocytic leukemia. Blood. 2018;131(14):1576-1586. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

