OpenMM 8: Molecular Dynamics Simulation with Machine Learning Potentials
- PMID: 38154096
- PMCID: PMC10846090
- DOI: 10.1021/acs.jpcb.3c06662
OpenMM 8: Molecular Dynamics Simulation with Machine Learning Potentials
Abstract
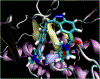
Machine learning plays an important and growing role in molecular simulation. The newest version of the OpenMM molecular dynamics toolkit introduces new features to support the use of machine learning potentials. Arbitrary PyTorch models can be added to a simulation and used to compute forces and energy. A higher-level interface allows users to easily model their molecules of interest with general purpose, pretrained potential functions. A collection of optimized CUDA kernels and custom PyTorch operations greatly improves the speed of simulations. We demonstrate these features in simulations of cyclin-dependent kinase 8 (CDK8) and the green fluorescent protein chromophore in water. Taken together, these features make it practical to use machine learning to improve the accuracy of simulations with only a modest increase in cost.
Figures
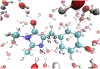
Update of
-
OpenMM 8: Molecular Dynamics Simulation with Machine Learning Potentials.ArXiv [Preprint]. 2023 Nov 29:arXiv:2310.03121v2. ArXiv. 2023. Update in: J Phys Chem B. 2024 Jan 11;128(1):109-116. doi: 10.1021/acs.jpcb.3c06662. PMID: 37986730 Free PMC article. Updated. Preprint.
References
-
- Thölke P; De Fabritiis G TorchMD-NET: Equivariant Transformers for Neural Network Based Molecular Potentials. arXiv April 23, 2022. 10.48550/arXiv.2202.02541. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

