Novel enhancer mediates the RPL36A-HNRNPH2 readthrough loci and GLA gene expressions associated with fabry disease
- PMID: 38155709
- PMCID: PMC10753776
- DOI: 10.3389/fgene.2023.1229088
Novel enhancer mediates the RPL36A-HNRNPH2 readthrough loci and GLA gene expressions associated with fabry disease
Abstract
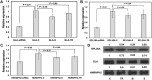
Fabry disease (FD) is a rare genetic condition caused by mutations in the GLA gene, located on the X chromosome in the RPL36-HNRNPH2 readthrough genomic region. This gene produces an enzyme called alpha-galactosidase A (α-Gal A). When the enzyme does not function properly due to the mutations, it causes harmful substances called globotriaosylceramide (Gb3) and globotriaosylsphingosine (lyso-Gb3) to build up in the body's lysosomes. This accumulation can damage the kidneys, heart, eyes, and nervous system. Recent studies have shown that the RPL36A-HNRNPH2 readthrough loci, which include RPL36A and HNRNPH2 genes, as well as the regulatory sequence known as the GLA-HNRNPH2 bidirectional promoter, may also play a role in FD. However, the involvement of enhancer RNAs (eRNAs) in FD is still poorly understood despite their known role in various diseases. To investigate this further, we studied an RPL36A enhancer called GH0XJ101390 and showed its genomic setting in the RPL36-HNRNPH2 readthrough region; the eRNA is rich in Homotypic Clusters of TFBSs (HCTs) type and hosts a CpG Island (CGI). To test the functional correlation further with GLA, RPL36A, and HNRNPH2, we used siRNAs to knock down GH0XJ101390 in human kidney embryonic cells 293T. The results showed a significant decrease in RPL36A and GLA expression and a non-significant decrease in HNRNPH2 expression. These findings could have important implications for understanding the regulatory mechanisms of GH0XJ101390 and its potential role in FD. A better understanding of these mechanisms may improve diagnostic and therapeutic methods for FD, which could ultimately benefit patients with this rare condition.
Keywords: bioinformatic; enhancer; fabry disease; lncRNA; readthrough locus; α-Gal A.
Copyright © 2023 Al-Obaide, Islam, Al-Obaidi and Vasylyeva.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Al-Obaide M. A., Al-Obaidi I. I., Vasylyeva T. L. (2021). Unexplored regulatory sequences of divergently paired GLA and HNRNPH2 loci pertinent to Fabry disease in human kidney and skin cells: presence of an active bidirectional promoter. Exp. Ther. Med. 21, 154. 10.3892/etm.2020.9586 - DOI - PMC - PubMed
-
- Al-Obaide M. A., Al-Obaidi I. I., Vasylyeva T. L. (2022). The potential consequences of bidirectional promoter methylation on GLA and HNRNPH2 expression in Fabry disease phenotypes in a family of patients carrying a GLA deletion variant. Biomed. Rep. 17, 71. 10.3892/br.2022.1554 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

