Signaling pathways of liver regeneration: Biological mechanisms and implications
- PMID: 38155779
- PMCID: PMC10753089
- DOI: 10.1016/j.isci.2023.108683
Signaling pathways of liver regeneration: Biological mechanisms and implications
Abstract
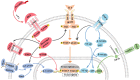
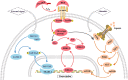
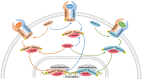
The liver possesses a unique regenerative ability to restore its original mass, in this regard, partial hepatectomy (PHx) and partial liver transplantation (PLTx) can be executed smoothly and safely, which has important implications for the treatment of liver disease. Liver regeneration (LR) can be the very complicated procedure that involves multiple cytokines and transcription factors that interact with each other to activate different signaling pathways. Activation of these pathways can drive the LR process, which can be divided into three stages, namely, the initiation, progression, and termination stages. Therefore, it is important to investigate the pathways involved in LR to elucidate the mechanism of LR. This study reviews the latest research on the key signaling pathways in the different stages of LR.
Keywords: Biological sciences; Health sciences.
© 2023 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Liver Regeneration after Hepatectomy and Partial Liver Transplantation.Int J Mol Sci. 2020 Nov 9;21(21):8414. doi: 10.3390/ijms21218414. Int J Mol Sci. 2020. PMID: 33182515 Free PMC article. Review.
-
iTRAQ-based quantitative proteomic analysis of the liver regeneration termination phase after partial hepatectomy in mice.J Proteomics. 2022 Sep 15;267:104688. doi: 10.1016/j.jprot.2022.104688. Epub 2022 Jul 29. J Proteomics. 2022. PMID: 35914716
-
Mitochondria-derived H2O2 triggers liver regeneration via FoxO3a signaling pathway after partial hepatectomy in mice.Cell Death Dis. 2023 Mar 28;14(3):216. doi: 10.1038/s41419-023-05744-w. Cell Death Dis. 2023. PMID: 36977674 Free PMC article.
-
Inhibition of wild-type p53-induced phosphatase 1 promotes liver regeneration in mice by direct activation of mammalian target of rapamycin.Hepatology. 2015 Jun;61(6):2030-41. doi: 10.1002/hep.27755. Epub 2015 Mar 25. Hepatology. 2015. PMID: 25704606
-
Biomechanics in liver regeneration after partial hepatectomy.Front Bioeng Biotechnol. 2023 May 5;11:1165651. doi: 10.3389/fbioe.2023.1165651. eCollection 2023. Front Bioeng Biotechnol. 2023. PMID: 37214300 Free PMC article. Review.
Cited by
-
Sequential activation of transcription factors promotes liver regeneration through specific and developmental enhancers.Cell Genom. 2025 Jul 9;5(7):100887. doi: 10.1016/j.xgen.2025.100887. Epub 2025 May 22. Cell Genom. 2025. PMID: 40409273 Free PMC article.
-
Multi-parametric atlas of the pre-metastatic liver for prediction of metastatic outcome in early-stage pancreatic cancer.Nat Med. 2024 Aug;30(8):2170-2180. doi: 10.1038/s41591-024-03075-7. Epub 2024 Jun 28. Nat Med. 2024. PMID: 38942992 Free PMC article.
-
Should I Stay or Should I Go - ERK as Control Lever in Hepatocyte Expansion.Cell Mol Gastroenterol Hepatol. 2025;19(9):101532. doi: 10.1016/j.jcmgh.2025.101532. Epub 2025 May 30. Cell Mol Gastroenterol Hepatol. 2025. PMID: 40456274 Free PMC article. No abstract available.
-
Zebrafish as a model for human epithelial pathology.Lab Anim Res. 2025 Feb 3;41(1):6. doi: 10.1186/s42826-025-00238-6. Lab Anim Res. 2025. PMID: 39901304 Free PMC article. Review.
-
Blood flow-induced angiocrine signals promote organ growth and regeneration.Bioessays. 2025 Feb;47(2):e2400207. doi: 10.1002/bies.202400207. Epub 2024 Nov 11. Bioessays. 2025. PMID: 39529434 Free PMC article. Review.
References
-
- Taki-Eldin A., Zhou L., Xie H.Y., Zheng S.S. Liver regeneration after liver transplantation. Eur. Surg. Res. 2012;48:139–153. - PubMed
-
- Pahlavan P.S., Feldmann R.E., Jr., Zavos C., Kountouras J. Prometheus' challenge: molecular, cellular and systemic aspects of liver regeneration. J. Surg. Res. 2006;134:238–251. - PubMed
-
- Fujiyoshi M., Ozaki M. Molecular mechanisms of liver regeneration and protection for treatment of liver dysfunction and diseases. J. Hepatobiliary. Pancreat. Sci. 2011;18:13–22. - PubMed
-
- Taub R. Liver regeneration: from myth to mechanism. Nat. Rev. Mol. Cell Biol. 2004;5:836–847. - PubMed
-
- Minuk G.Y. Hepatic regeneration: If it ain't broke, don't fix it. Can. J. Gastroenterol. 2003;17:418–424. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

