Preclinical immune efficacy against SARS-CoV-2 beta B.1.351 variant by MVA-based vaccine candidates
- PMID: 38155964
- PMCID: PMC10754519
- DOI: 10.3389/fimmu.2023.1264323
Preclinical immune efficacy against SARS-CoV-2 beta B.1.351 variant by MVA-based vaccine candidates
Abstract
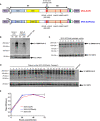
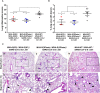
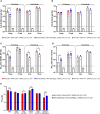
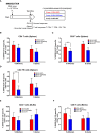
The constant appearance of new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VoCs) has jeopardized the protective capacity of approved vaccines against coronavirus disease-19 (COVID-19). For this reason, the generation of new vaccine candidates adapted to the emerging VoCs is of special importance. Here, we developed an optimized COVID-19 vaccine candidate using the modified vaccinia virus Ankara (MVA) vector to express a full-length prefusion-stabilized SARS-CoV-2 spike (S) protein, containing 3 proline (3P) substitutions in the S protein derived from the beta (B.1.351) variant, termed MVA-S(3Pbeta). Preclinical evaluation of MVA-S(3Pbeta) in head-to-head comparison to the previously generated MVA-S(3P) vaccine candidate, expressing a full-length prefusion-stabilized Wuhan S protein (with also 3P substitutions), demonstrated that two intramuscular doses of both vaccine candidates fully protected transgenic K18-hACE2 mice from a lethal challenge with SARS-CoV-2 beta variant, reducing mRNA and infectious viral loads in the lungs and in bronchoalveolar lavages, decreasing lung histopathological lesions and levels of proinflammatory cytokines in the lungs. Vaccination also elicited high titers of anti-S Th1-biased IgGs and neutralizing antibodies against ancestral SARS-CoV-2 Wuhan strain and VoCs alpha, beta, gamma, delta, and omicron. In addition, similar systemic and local SARS-CoV-2 S-specific CD4+ and CD8+ T-cell immune responses were elicited by both vaccine candidates after a single intranasal immunization in C57BL/6 mice. These preclinical data support clinical evaluation of MVA-S(3Pbeta) and MVA-S(3P), to explore whether they can diversify and potentially increase recognition and protection of SARS-CoV-2 VoCs.
Keywords: COVID-19; MVA-based vaccine; S protein; SARS-CoV-2; efficacy; immunogenicity; mice; variants of concern.
Copyright © 2023 Pérez, Albericio, Astorgano, Flores, Sánchez-Corzo, Sánchez-Cordón, Luczkowiak, Delgado, Casasnovas, Esteban and García-Arriaza.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
MVA-based vaccine candidates expressing SARS-CoV-2 prefusion-stabilized spike proteins of the Wuhan, Beta or Omicron BA.1 variants protect transgenic K18-hACE2 mice against Omicron infection and elicit robust and broad specific humoral and cellular immune responses.Front Immunol. 2024 Aug 29;15:1420304. doi: 10.3389/fimmu.2024.1420304. eCollection 2024. Front Immunol. 2024. PMID: 39267752 Free PMC article.
-
Intranasal administration of a single dose of MVA-based vaccine candidates against COVID-19 induced local and systemic immune responses and protects mice from a lethal SARS-CoV-2 infection.Front Immunol. 2022 Sep 12;13:995235. doi: 10.3389/fimmu.2022.995235. eCollection 2022. Front Immunol. 2022. PMID: 36172368 Free PMC article.
-
A Single Dose of an MVA Vaccine Expressing a Prefusion-Stabilized SARS-CoV-2 Spike Protein Neutralizes Variants of Concern and Protects Mice From a Lethal SARS-CoV-2 Infection.Front Immunol. 2022 Jan 27;12:824728. doi: 10.3389/fimmu.2021.824728. eCollection 2021. Front Immunol. 2022. PMID: 35154086 Free PMC article.
-
COVID-19 Pandemic and Vaccines Update on Challenges and Resolutions.Front Cell Infect Microbiol. 2021 Sep 10;11:690621. doi: 10.3389/fcimb.2021.690621. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34568087 Free PMC article. Review.
-
Immunology and Technology of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Vaccines.Pharmacol Rev. 2022 Jan;74(1):313-339. doi: 10.1124/pharmrev.120.000285. Pharmacol Rev. 2022. PMID: 35101964 Review.
Cited by
-
Design and construction of a fast synthetic modified vaccinia virus Ankara reverse genetics system for advancing vaccine development.Front Microbiol. 2025 Apr 25;16:1572706. doi: 10.3389/fmicb.2025.1572706. eCollection 2025. Front Microbiol. 2025. PMID: 40351316 Free PMC article.
-
Specific Immune Responses and Oncolytic Effects Induced by EBV LMP2A-Armed Modified Ankara-Vaccinia Virus Vectored Vaccines in Nasopharyngeal Cancer.Pharmaceutics. 2025 Jan 3;17(1):52. doi: 10.3390/pharmaceutics17010052. Pharmaceutics. 2025. PMID: 39861700 Free PMC article.
-
MVA-based vaccine candidates expressing SARS-CoV-2 prefusion-stabilized spike proteins of the Wuhan, Beta or Omicron BA.1 variants protect transgenic K18-hACE2 mice against Omicron infection and elicit robust and broad specific humoral and cellular immune responses.Front Immunol. 2024 Aug 29;15:1420304. doi: 10.3389/fimmu.2024.1420304. eCollection 2024. Front Immunol. 2024. PMID: 39267752 Free PMC article.
-
Comprehensive review of preclinical evaluation strategies for COVID-19 vaccine candidates: assessing immunogenicity, toxicology, and safety profiles.Iran J Microbiol. 2025 Feb;17(1):1-18. doi: 10.18502/ijm.v17i1.17796. Iran J Microbiol. 2025. PMID: 40330066 Free PMC article. Review.
-
Implementation of an Immunoassay Based on the MVA-T7pol-Expression System for Rapid Identification of Immunogenic SARS-CoV-2 Antigens: A Proof-of-Concept Study.Int J Mol Sci. 2024 Oct 10;25(20):10898. doi: 10.3390/ijms252010898. Int J Mol Sci. 2024. PMID: 39456680 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

